FIGURE 3.
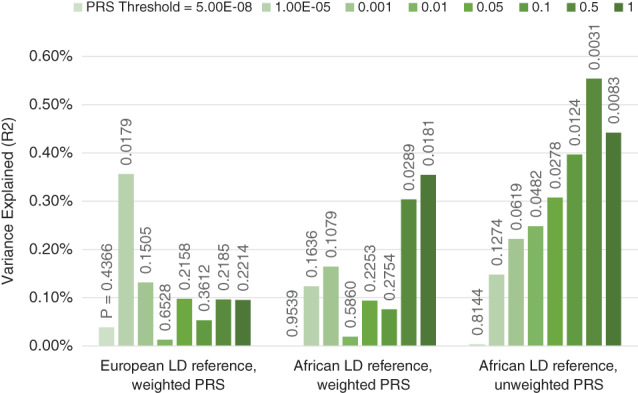
Performance of cross‐ancestry polygenic score prediction as a function of LD reference data and weighting. PRS analyses were conducted using 966 unrelated participants of African ancestry from the ABCD study (Casey et al., 2018) (relatedness <.025; ancestry defined as <6 SD from the African reference centroid in a multidimensional scaling analysis). The polygenic scores were derived from a European ancestry meta‐analysis of the hippocampal volume GWAS from Hibar et al., 2015 and a GWAS of 20,111 individuals from the UK Biobank. PRS were calculated using the traditional clumping and thresholding approach (‐‐clump‐r2 0.1 ‐‐clump‐kb 1,000) using reference sets of 1,000 participants of European or African ancestry (sampled from the ABCD study). For the weighted PRS, the SNP dosages were multiplied by the betas from the GWAS as is traditionally done. For the unweighted PRS the betas from the GWAS were recoded to 1 if β > 0, −1 if β < 0 and 0 if β = 0. Analyses included Age, Sex, ancestry corrections (components 1–4) and ICV as covariates
