Abstract
Background
Biofilms are a main pathogenicity feature of Pseudomonas aeruginosa and has a significant role in antibiotic resistance and persistent infections in humans. We investigated the in vitro activities of antibiotic ceftazidime and enzyme cellulase, either alone or in combination against biofilms of P. aeruginosa.
Results
Both ceftazidime and cellulase significantly decreased biofilm formation in all strains in a dose-dependent manner. Combination of enzyme at concentrations of 1.25, 2.5, 5, and 10 U/mL tested with 1/16× MIC of antibiotic led to a significant reduction in biofilm biomass. Cellulase showed a significant detachment effect on biofilms at three concentrations of 10 U/mL, 5 U/mL, and 2.5 U/mL. The MIC, MBC, and MBEC values of ceftazidime were 2 to 4 µg/mL, 4 to 8 µg/mL, and 2048 to 8192 µg/mL. When combined with cellulase, the MBECs of antibiotic showed a significant decrease from 32- to 128-fold.
Conclusions
Combination of the ceftazidime and the cellulase had significant anti-biofilm effects, including inhibition of biofilm formation and biofilm eradication in P. aeruginosa. These data suggest that glycoside hydrolase therapy as a novel strategy has the potential to enhance the efficacy of antibiotics and helps to resolve biofilm-associated wound infections caused by this pathogen.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-021-02411-y.
Keywords: Biofilm inhibition, Ceftazidime, Cellulase, MBEC, Pseudomonas aeruginosa
Background
Biofilms are communities of microorganisms protected by a self-synthesized layer of complex polymers, called the extracellular polymeric substances (EPSs), represented mainly by proteins (>2%), polysaccharides (1–2%), and other constituents, such as extracellular DNA (eDNA) (<1%), RNA (<1%), and bound and free ions [1, 2]. Biofilms may form on biotic or abiotic surfaces and be prevalent in natural, industrial, and hospital settings. The microbial cells growing in a biofilm are physiologically distinct from planktonic cells of the same organism, which, by contrast, are single-cells that may float or swim in a liquid medium [3].
The biofilm mediates bacterial stability and protects them from environmental conditions, such as pH, UV damage, hydrogen peroxide, metal, and antibiotic pressure [4]. Likewise, the biofilm’s exopolysaccharide component acts as a barrier against the host immune response towards bacterial infection, including phagocytosis [5]. It has also been known that biofilm-forming bacteria are up to 1000-times more resistant to antimicrobial agents than planktonic cells, partially due to the impaired penetration [6]. Multiple mechanisms of biofilm formation and its architectural features, including decreased transportation and diffusion of antimicrobial agents into the extracellular matrix supported by glycocalyx matrix, heterogeneity in metabolism and growth rate, increased activity of multidrug efflux pumps, the formation of antimicrobial tolerant-persisters and slow-growing cells, genetic adaptation, stress responses, and quorum-sensing systems confer the increased resistance in biofilm-forming bacteria [6, 7].
According to the National Institutes of Health (NIH) agency, bacterial biofilms are involved in 65% of microbial diseases and more than 80% of chronic infections [8]. One bacterium that is particularly notorious for forming biofilms is Pseudomonas aeruginosa [9–11]. P. aeruginosa is a major nosocomial pathogen associated with a wide variety of acute and chronic infections, such as soft tissue infections, urinary tract infections, bacteremia, pneumonia, diabetic foot, otitis, and keratitis [12]. Burn wound infections are one of the most important complications following the burn injuries and may be associated with increased morbidity and mortality [13, 14]. The ability of P. aeruginosa to form biofilms is considered one of the main pathogenicity features of this organism in causing persistent infections in such cases, resulting in delayed healing for 2 to 4 weeks [15, 16].
Given the intrinsic resistance of P. aeruginosa biofilm to antimicrobial therapy, researchers are trying to develop new strategies that explicitly target established biofilms, hoping that it can significantly improve clinical practice [6]. An alternate anti-biofilm approach is the use of therapeutic enzymes that degrade the biofilm matrix. An enzyme complex (amylase, cellulase, and protease) was found that was able to degrade the biofilms of different bacteria, including Escherichia coli (85.5%), Salmonella enterica (79.72%), P. aeruginosa (88.76%), and Staphyloccus aureus (87.42%) [17]. Glycoside hydrolases (also called glycosidases) hydrolyze glycosidic bonds in complex carbohydrates [18]. Cellulase is a type of glycoside hydrolase that acts specifically by breaking down the β-1,4 linkages in polysaccharides, such as cellulose, an exopolysaccharide commonly found in the biofilm of several bacteria, including E. coli, Salmonella, Citrobacter, Enterobacter, and Pseudomonas as well as Agrobacterium tumefaciens [19]. As exopolysaccharides represent a substantial component of many bacterial biofilms and contribute to the structural stability of the EPS [20], their disruption is essential for dispersing bacteria into a planktonic state. This would increase the host immune system’s availability and administered antimicrobial compounds/antibiotics to infected cells. So, it is thought that hydrolyzing the glycosidic linkages that hold exopolysaccharides together will degrade the EPS. It has been shown that cellulase inhibits the growth of biofilms produced by Burkholderia cepacia and P. aeruginosa on various abiotic surfaces that are commonly used in medical devices [21, 22].
In this study, we aimed to determine the in vitro activities of anti-pseudomonal antibiotic, ceftazidime, and glycoside hydrolase enzyme, cellulase, alone or in combination against biofilms of standard and clinical isolates of P. aeruginosa.
Methods
Chemicals
Ceftazidime hydrate (CAS# 120618-65-7) and fungal cellulase (from Aspergillus niger) (CAS# 9012-54-8) were purchased from Sigma-Aldrich (St Louis, MO, USA). Ceftazidime stock solution was prepared by dissolving the lyophilized powder in sodium carbonate buffer. Cellulase stock solution was prepared by dissolving the dry powder in sodium acetate buffer (50 mM, pH 5) at 65 °C for 5 min. The heat-inactivated enzyme was obtained by heating the enzyme solution at 95 °C for 20 min. All stock solutions were stored at −80 °C freezer for up to 6 months.
Culture media
Mueller–Hinton broth (MHB) was used for the determination of minimum inhibitory concentration (MIC) values and the microbroth chequerboard technique, tryptic soy broth supplemented with 1% glucose (TSB-glucose) used for biofilm production and inhibition assays, and tryptic soy agar was used for determination of the minimum bactericidal concentration (MBC) were obtained from Merck (Merck KGaA, Darmstadt, Germany).
Bacterial strains
The bacterial strains used in all experiments were as follows: P. aeruginosa wild-type strain PAO1, a laboratory reference strain originally isolated in 1954 from an infected burn/wound of a patient in Melbourne, Australia (American Type Culture Collection ATCC 15,692) and two clinical isolates of P. aeruginosa recovered from the patients with burn wound infections that were identified by standard microbiological and biochemical methods from a previous study [9].
Antibacterial activity assays
The MIC of ceftazidime was determined by the microbroth dilution method in 96-well polystyrene microplates (JET Biofil, Guangzhou, China) according to the Clinical and Laboratory Standards Institute (CLSI) guideline [23]. The MIC was defined as the lowest concentration of antibiotic that completely inhibited visible growth. The MBC value was determined at the end of the incubation period by removing 10 µL aliquots of all wells with no visible growth and dispensed in tryptic soy agar. Grown colonies were counted after 48 h incubation at 37 °C. The MBC was established as the lowest concentration of antibiotic that killed at least 99.9% of the initial inoculums.
The bactericidal activity of cellulase on planktonic cells of P. aeruginosa was investigated. Briefly, the overnight cultures were treated with two-fold serial dilutions of enzyme ranging from 0.09 U/mL to 50 U/mL in the wells of polystyrene microplate as described for the MIC determination. A well containing bacteria exposed with heat-inactivated enzyme was used as control. After 24 h incubation at 37 °C, the effect of the enzyme on the bacterial growth was determined by measuring the optical density (OD) of each culture in wells at 570 nm.
Biofilm formation assay
Biofilm formation was assessed by the colorimetric microtiter plate assay as described previously [9]. An overnight culture of P. aeruginosa was adjusted to the turbidity of a 0.5 McFarland standard and then was diluted 1:100 in TSB-glucose, yielding a final concentration of about 1 × 106 CFU/200 µL. This suspension was added to each well of a sterile 96-well polystyrene microplate and incubated at 37 °C for 24 h. Then, the supernatant was carefully aspirated from the wells followed by washing three times with 200 µL sterile phosphate-buffered saline (PBS, pH 7.3). Adherent biofilm in each well was fixed by 99% methanol for 15 min, the solutions were removed, and the plate was allowed to dry. Wells were stained with 200 µL of 0.1% crystal violet (CV) (Sigma Chemical Co., St Louis, MO, USA) for 5 min at room temperature, rinsed by water, and then allowed to dry. The stain was dissolved with 200 µL of 95% ethanol solution for 30 min. The OD of each well was determined at 595 nm in a microtiter plate reader (BioTek, Bad Friedrichshall, Germany). All experiments were evaluated in triplicate. A cut-off value (ODc) as three standard deviations (SD) above the mean OD of the negative control was established: ODc = average OD of negative control + (3 × SD of negative control). The isolates were divided into four categories: non-biofilm producer (OD < ODc), weak-biofilm producer (ODc < OD < 2 × ODc), moderate-biofilm producer (2 × ODc < OD < 4 × ODc), and strong-biofilm producer (4 × ODc < OD).
Biofilm attachment assay
Biofilm attachment assays were performed using a previously described method with some modifications [24]. The overnight cultures were diluted 1:100 to give 1 × 106 CFU/200 µL in TSB-glucose, and standard and clinical P. aeruginosa strains were added to each well of a 96-well microtitre plate with 1× and 1/2× MIC concentrations of ceftazidime or 1.25, 2.5, 5, and 10 U/mL of cellulase. The plates were incubated for 1, 2, or 4 h at 37 °C. The positive controls were P. aeruginosa strains in TSB-glucose without antibiotic or enzyme, and negative controls were medium with neither bacteria nor antimicrobial agents. After incubation, the wells were washed three times with PBS, and biofilm biomass was assessed by CV staining as described above.
Inhibition of biofilm formation
Bacterial strains were prepared at a concentration of 1 × 106 CFU/200 µL in TSB-glucose were added to each well of a microtitre plate with 1×, 1/2×, 1/4×, 1/8×, and 1/16× MIC concentrations of ceftazidime, 1.25, 2.5, 5, and 10 U/mL of cellulase, and combination. Wells inoculated with bacterial strains without antibiotic or enzyme were set up as positive controls. At the end of the 24-hour incubation period, wells were rinsed three times with PBS, and biofilm biomass was assessed by CV staining. The results expressed as the percentage biofilm biomass reduction compared with wells without antibiotic or enzyme.
Biofilm treatment assay by the enzyme
The efficacy of cellulase in affecting biofilm detachment was determined. Briefly, the established biofilms in a microtiter plate were washed three times with PBS and exposed to different enzyme concentrations (range 0.01 to 10 U/mL). TSB-glucose without enzyme or bacteria was used as negative control, and biofilm with no enzyme treatment was considered positive control. A well containing bacteria treated with heat-inactivated (HI) enzyme was also used as control. After 1-h incubation at 37 °C, well contents were removed and washed thrice with PBS. Biofilm in each well was stained using the CV assay, and OD was determined at 595 nm.
Minimum biofilm eradication concentration (MBEC) assay
Antimicrobial susceptibilities of standard and clinical P. aeruginosa biofilms were evaluated according to the method of Naparstek et al. [25] with few modifications. The mature biofilms in a 96-well microtiter plate were washed three times with PBS to remove planktonic bacteria. Serial 2-fold dilutions of ceftazidime ranging from 64 to 32,768 µg/mL were prepared in cation-adjusted Mueller-Hinton broth (CAMHB). A 200 µL sample of each concentration was added to a corresponding well, and plates were incubated overnight at 37 °C. The wells with established biofilm and CAMHB medium, without antibiotic and enzyme treatment were considered positive and negative controls, respectively. After the incubation, plates were washed three times with sterile PBS to remove residual antibiotic, and 200 µL of fresh CAMHB was placed in each well for further 24 h incubation at 37 °C. This allows the biofilm bacteria that survived the antibiotic exposure to grow in the absence of antibiotic and produce detectable turbidity. Subsequently, the OD of each well was determined at 595 nm. The lowest antibiotic concentration that prevented bacterial regrowth from the treated biofilm was defined as the MBEC value.
The effect of cellulase on ceftazidime MBEC
The combined effect of cellulase and ceftazidime on P. aeruginosa biofilms was determined as described previously [26, 27]. Briefly, bacterial biofilms were exposed to 2-fold diluted ceftazidime concentrations (range 2 to 1024 µg/mL) and various concentrations of cellulase (1.25, 2.5, 5, and 10 U/mL). Following 24-h incubation, the well content was removed and washed three times with PBS, and then fresh CAMHB was added to each well for further overnight incubation. The MBEC of ceftazidime for biofilm cultures was as described above in “MBEC assay”.
Statistical analysis
All experiments were performed in three independent assays. The results were expressed as means ± standard deviations of three independent experiments for the biofilm attachment and inhibition of biofilm formation assays. A one-way analysis of variance (ANOVA) followed by Tukey-Kramer multiple comparison tests was used to evaluate differences between groups’ mean values. A P-value < 0.05 was considered statistically significant. All tests were performed using GraphPad statistical software (GraphPad Software, San Diego, CA, USA).
Results
Susceptibility
In general, P. aeruginosa PAO1 strain and two clinical isolates presented low MICs to ceftazidime, in the range of the susceptible category (2-4 µg/mL) (Table 1). The MBC values were also two-fold greater than those of the MIC values.
Table 1.
In vitro antibacterial and anti-biofilm activities of ceftazidime against P. aeruginosa strains
| aBacterial strain | MIC (µg/mL) | MBC (µg/mL) | MBEC (µg/mL) | Fold change in MBEC/MIC ratio | Fold change in MBEC/MBC ratio |
|---|---|---|---|---|---|
| PAO1 | 2 | 4 | 2048 | 1024 | 512 |
| PA1 | 4 | 8 | 4096 | 1024 | 512 |
| PA2 | 2 | 4 | 8192 | 4096 | 2048 |
MBEC Minimum biofilm eradication concentration, MIC Minimum inhibitory concentration, MBC Minimum bactericidal concentration
a PAO1 is a laboratory reference strain of P. aeruginosa and PA1 and PA2 are two clinical isolates of P. aeruginosa that were recovered from the patients with burn wound infections
The bactericidal activity of cellulase on P. aeruginosa planktonic cells was investigated. The enzyme had no bactericidal effect, and bacteria grew after treatment with all concentrations (0.09-50 U/mL) of this enzyme (P > 0.05).
Biofilm formation
The results of biofilm formation assay performed on three P. aeruginosa strains showed strong-mass biofilms (OD595 range 2.55–3.28) (P > 0.05). The isolate PA1 was found to produce more biofilm than the standard strain PAO1 or the isolate PA2.
Biofilm attachment
Figure 1 shows the effects of ceftazidime at MIC, and 1/2× MIC concentrations on the adherence of P. aeruginosa strains to the wells of microtitre plate after 1, 2, or 4 h incubation at 37 °C. In two P. aeruginosa clinical isolates PA1 and PA2, none of the concentrations used inhibited biofilm attachment for each incubation period (Fig. 1B, C), while antibiotic achieved this inhibition in PAO1 strain in a concentration- and time-dependent manner; the MIC and 1/2× MIC concentrations inhibited 64% and 57% of attachment, respectively, after four h of incubation (Fig. 1 A). Besides, cellulase did not affect the attachment of P. aeruginosa at concentrations of 1.25, 2.5, 5, and 10 U/mL tested (Data are not shown).
Fig. 1.
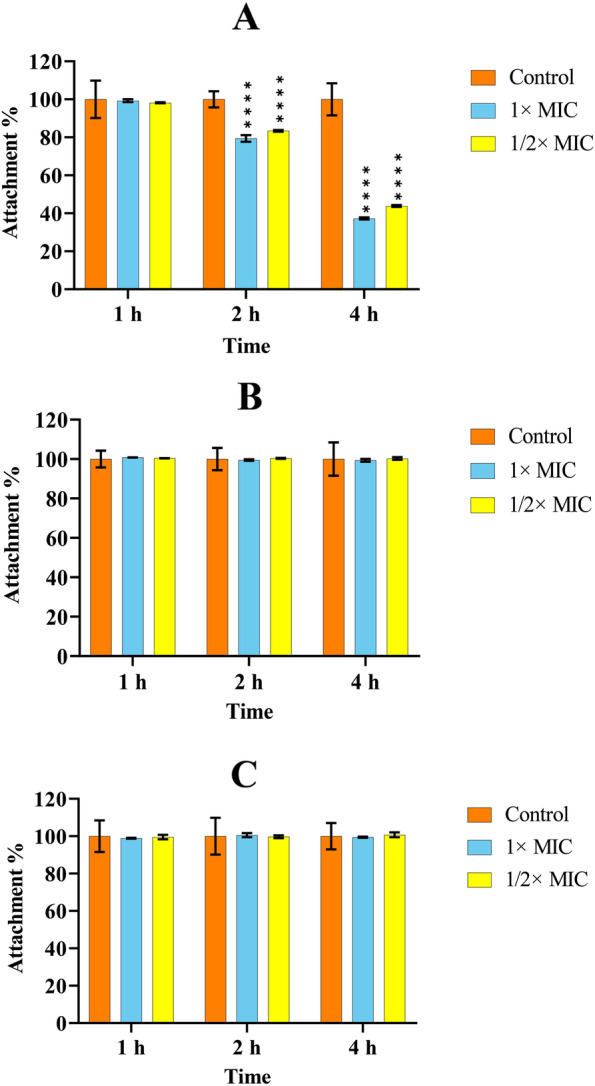
The results of the P. aeruginosa attachment to the surface of microplate wells containing 1× MIC and 1/2× MIC concentrations of ceftazidime in PAO1 (A), PA1 (B), and PA2 (C). The plates were incubated for 1, 2, or 4 h at 37 °C. Data were normalized to the mean value of the control, which was set at 100%. The error bars indicate the standard deviations between bacteria. Results were expressed as percentage of biofilm formed with respect to control. The statistical significance of the data was determined by an analysis of variance (ANOVA) test followed by the Tukey-Kramer multiple comparison test. Significance was accepted when the P-value was < 0.05 (****P < 0.0001). MIC: Minimum inhibitory concentration
Inhibition of biofilm formation
The ability of ceftazidime and cellulase at different concentrations to inhibit biofilm formation was evaluated by CV staining after a 24 h-incubation of P. aeruginosa strains. As shown in Fig. 2, both antibiotic and enzyme significantly decreased biofilm formation in all strains in a dose-dependent manner compared to cells incubated in medium only (P < 0.0001). There was only moderate biofilm inhibition when all P. aeruginosa strains were incubated with ceftazidime at a concentration of 1/16× MIC (Fig. 2 A). Likewise, the 1.25 U/mL concentration of cellulase showed minor activity in the production of biofilm in all P. aeruginosa strains (Fig. 2B).
Fig. 2.
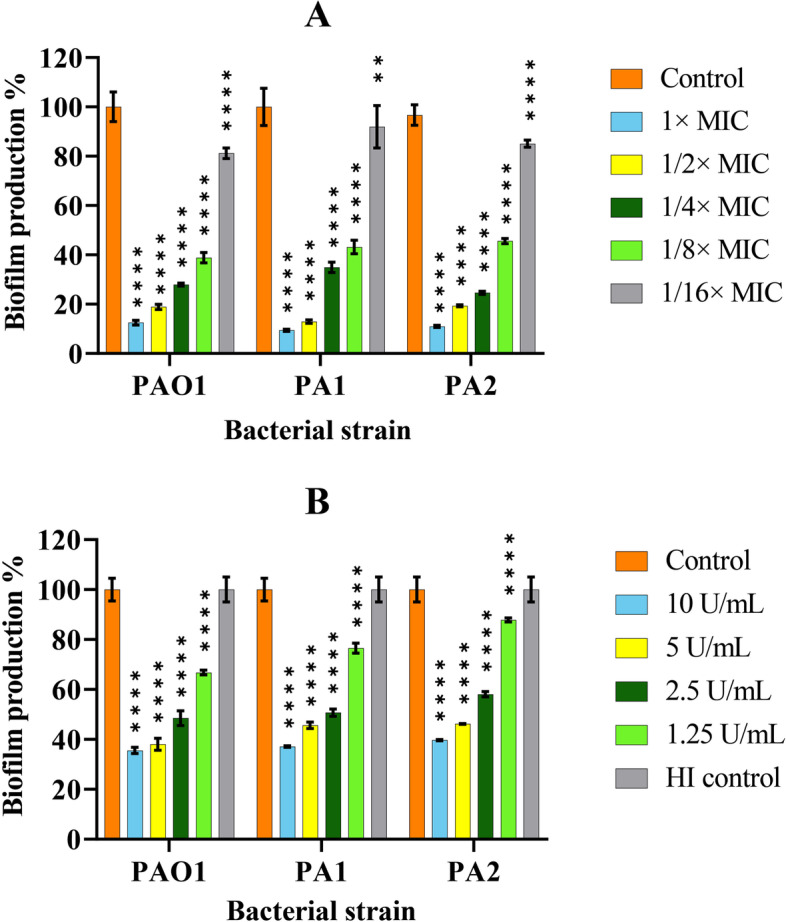
Reduction in the P. aeruginosa biofilm formation by PAO1, PA1, and PA2 with different concentrations of ceftazidime (A) and cellulase (B). The error bars indicate the standard deviations between strains. The microplates were incubated for 24 h at 37 °C. Data were normalized to the mean value of the control, which was set at 100%. The error bars indicate the standard deviations between bacteria. Results were expressed as percentage of biofilm biomass formed with respect to control. The statistical significance of the data was determined by an analysis of variance (ANOVA) test followed by the Tukey-Kramer multiple comparison test. Significance was accepted when the P-value was < 0.05 (****P < 0.0001, **P < 0.01). HI: Heat-inactivated; MIC: Minimum inhibitory concentration
Biofilm treatment with enzyme
The biofilm detachment activity of serial dilutions of cellulase against P. aeruginosa is shown in Table 2. Cellulase showed a significant detachment effect on biofilms at three concentrations of 10 U/mL, 5 U/mL, and 2.5 U/mL, with a biofilm reduction of 77.3-78.6%, 72.1-73.1%, and 78.3-78.4% in PAO1, PA1, and PA2, respectively, compared to the untreated control biofilms (P < 0.0001) (Data are also shown in supplementary Fig. 1). In addition, no reduction in biofilm biomass was observed with the HI enzyme.
Table 2.
The effects of serial dilutions of cellulase on reduction of mature biofilms of P. aeruginosa strains
| aBacterial strain | % of biofilm forming reduction by cellulase concentration (U/mL) of | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.01 | 0.03 | 0.07 | 0.15 | 0.31 | 0.62 | 1.25 | 2.5 | 5 | 10 | No/inactivated enzyme | |
| PAO1 | 0.2 | 0.2 | 0.2 | 0.3 | 0.2 | 3 | 26.3 | 76.2 | 78.6 | 77.3 | 0 |
| PA1 | 0.2 | 0.2 | 0.3 | 0.4 | 0.3 | 2.9 | 10.8 | 72.1 | 72.7 | 73.1 | 0 |
| PA2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.5 | 3.9 | 17.4 | 78.4 | 77.9 | 78.3 | 0 |
a PAO1 is a laboratory reference strain of P. aeruginosa and PA1 and PA2 are two clinical isolates of P. aeruginosa that were recovered from the patients with burn wound infections
MBECs of ceftazidime
The MBEC results for bacterial biofilm are listed in Table 1. Data indicated that all three isolates, which are ceftazidime-susceptible in the planktonic state, showed a dramatic increase in resistance to ceftazidime when grown in biofilms (P < 0.0001); the ceftazidime MBEC was 1024 to 4096-fold higher (range 2048 to 8192 µg/mL) compared with the MIC (range 2 to 4 µg/mL). Likewise, compared with the MBC value (range 4 to 8 µg/mL), the MBEC was 512 to 2048-fold higher (range 512 to 2048 µg/mL).
Combination effect of cellulase and ceftazidime on biofilm formation
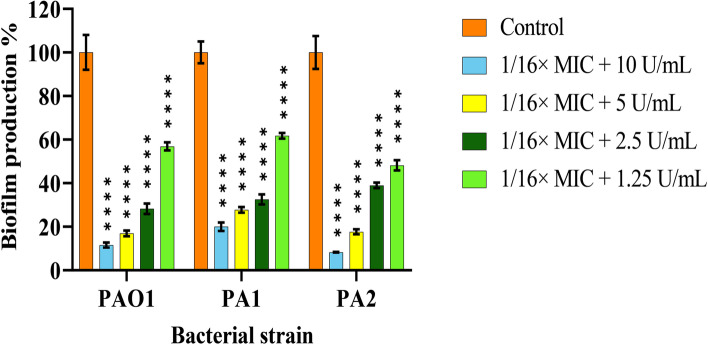
The sub-inhibitory concentration of 1/16× MIC of ceftazidime was chosen in combination with different concentrations of cellulase to assess the inhibitory effect of biofilm formation. Combination of the enzyme at all concentrations used with 1/16× MIC of antibiotic led to biofilm biomass reduction statistically higher than that caused by cellulase or ceftazidime used at the same concentrations alone (Fig. 3).
Fig. 3.
Reduction of P. aeruginosa biofilm formation by combination of ceftazidime (1/16× MIC) and different concentrations of cellulase in PAO1, PA1, and PA2. The error bars indicate the standard deviations between strains. The microplates were incubated for 24 h at 37 °C. Data were normalized to the mean value of the control, which was set at 100%. The error bars indicate the standard deviations between bacteria. Results were expressed as percentage of biofilm biomass formed with respect to control. The statistical significance of the data was determined by an analysis of variance (ANOVA) test followed by the Tukey-Kramer multiple comparison test. Significance was accepted when the P-value was < 0.05 (****P < 0.0001). MIC: Minimum inhibitory concentration
The effect of cellulase on ceftazidime MBEC
The combination of cellulase at all concentrations of 2.5, 5, and 10 U/mL and ceftazidime significantly decreased the ceftazidime MBECs from 32- to 128-fold (P < 0.05) (Table 3). In particular, the enzyme significantly reduced the ceftazidime MBEC in the PAO1 strain up to 128 times.
Table 3.
The combined effects of cellulase and ceftazidime on MBEC values of ceftazidime in P. aeruginosa strains
| Bacteria | MBEC value (µg/mL) of | Fold reduction in ceftazidime MBEC in the presence of cellulase | |
|---|---|---|---|
| Ceftazidime | Ceftazidime + cellulase | ||
| PAO1 | 2048 | 16 | 128 |
| PA1 | 4096 | 64 | 64 |
| PA2 | 8192 | 256 | 32 |
MBEC Minimum biofilm eradication concentration
Discussion
Generally, biofilm-associated infections are complicated to successfully treat due to high resistance to antibiotics [28, 29]. To overcome biofilm-associated resistance, new therapeutic approaches have been of interest to the scientific community and are being evaluated. In recent years, enzymes have been considered as potential anti-biofilm agents. Different types of enzymes have been used as anti-biofilm agents in treating infections or wound debridement [30–34]. Several studies have also demonstrated the ability of exogenous glycoside hydrolases to inhibit biofilm formation and degrade preexisting biofilms [1, 17]. Baker et al. have recently shown that Pel- and Psl-specific glycoside hydrolases, PelAh and PslGh, can inhibit the formation of and degrade biofilms of P. aeruginosa in vitro [35].
In this study, we investigated the in vitro activities of anti-pseudomonal antibiotic, ceftazidime, and the glycoside hydrolase enzyme, cellulase, alone and in combination against P. aeruginosa planktonic cells and biofilms. Results showed that cellulase had no bactericidal effect at the concentrations tested, consistent with those obtained in other studies that tested different enzymes [1, 35, 36]. Also, the destructive activity of various enzymes, including cellulase, on bacterial biofilms has been shown [1, 17, 37–39]. Two separate studies by Fleming et al. illustrated these results well. One study demonstrated that a dual-enzyme combination of α-amylase and cellulase don’t have any bactericidal activity but can significantly reduce the biofilm biomass of Staphylococcus aureus and P. aeruginosa, grown in vitro and in vivo, leading to the cell dispersal [1]. The second study showed that treatment of 48-hour-old S. aureus and P. aeruginosa mouse wound infections with 10% GH, α-amylase, and cellulase resulted in dispersal and significant septicemia, whereas pre-treating with 10% GH before inoculation did not cause an increase in septicemia, indicating that the effect of GH on bacteremia is directly on the biofilm [40]. Nagraj et al. showed an enzyme complex containing amylase, cellulase, and protease degraded biofilm of P. aeruginosa as high as 88.76% within one h of incubation [17]. Furthermore, a recent study demonstrated that the use of cellulase in multi-well plate biofilm dispersal assays leads to increased percent dispersal of Entrococcus faecalis [41]. Collectively, the biofilm degradation of bacteria by glycoside hydrolases, such as cellulase may be due to the hydrolysis of polysaccharides and disruption of the biofilm architecture that increased dispersal of planktonic cells. As a result, an increased rate of planktonic cell killing occurs by both the immune system and antimicrobial agents [6, 42] due partly to the higher metabolic rates and better cell surface access to free-floating cells.
When we examined the MIC values of ceftazidime against planktonic P. aeruginosa cells, they ranged between 2 and 4 µg/mL. When we considered the anti-biofilm activities of this antibiotic, MBEC values ranged between 2084 and 8192 µg/mL. According to these results, the MBEC/MIC ratio of this active antibiotic was found to be within the range of 1024- to 4096-fold. These results are similar to those obtained by others [43, 44].
The bacterial adhesion to surfaces is an essential step for biofilm formation. Many continuing studies target the inhibition of this critical step. In this study, we investigated the inhibition of bacterial attachment to the surfaces and the inhibition of biofilm production by MIC or subMIC values of ceftazidime or different concentrations of cellulase. Ceftazidime inhibited the attachment of PAO1 strain in a concentration- and time-dependent manner (P < 0.0001). Similar results were reported by Dosler et al. [43], where different antimicrobial agents, including ceftazidime at 1/10× MIC inhibited the biofilm attachment in a time-dependent fashion. The time-dependent killing of ceftazidime on planktonic P. aeruginosa cells, as demonstrated by Hengzhuang et al. [45], might explain no inhibitory effect of antibiotic on biofilm attachment observed in two clinical isolates. Neither of the tested concentrations of cellulase inhibited biofilm attachment in all bacteria, in accordance with results obtained by Cardeiro and coworkers [46]. Their results showed that subtilisin A reduced the attachment of P. aeruginosa by 44% onto a copolymer film, but cellulase had no significant effect, suggesting a more prominent role of proteins than polysaccharides in the initial attachment of P. aeruginosa to surfaces [46].
Our results also showed that ceftazidime and cellulase significantly inhibit biofilm formation in a concentration-manner, especially at 1× or 1/2× MIC (P < 0.0001). Similarly, Otani et al. observed a dose-dependent manner of reduction in biofilm formation by PAO1 strain with different concentrations (0, 1/16, 1/8, 1/4, 1/2, 1× MIC) of ceftazidime [47]. Given the bactericidal mechanism of ceftazidime, this anti-biofilm activity can be attributed to reducing bacterial density in a microbial population. Besides, downregulation of the pelA and pslA genes by subMIC ceftazidime has been shown by Otani et al. [47], suggesting a mechanism responsible for inhibited biofilm formation in P. aeruginosa. Likewise, significant reduction of biofilm formation at different cellulase concentrations indicates that enzyme targets polysaccharides playing during biofilm development and maturation in P. aeruginosa.
Moreover, our data demonstrated that ceftazdidme/cellulase combination significantly decreased biofilm formation compared to when agents were used at the same concentrations alone (P < 0.0001). The increased susceptibility of P. aeruginosa to various antibiotics in combination with PelAh and PslGh enzymes has been reported in two individual studies by Baker and colleagues [35, 48]. They demonstrated that treatment of PAO1 biofilms in vitro with these GH enzymes improves the efficacy of tobramycin, polymyxin, colistin, and neomycin, leading to nearly 1 log greater bacterial killing than antibiotic treatment alone [48]. Moreover, PelAh and PslGh treatment [35] and PslGh treatment [48] improves colistin and tobramycin penetration into the P. aeruginosa biofilm, leading to reduced viable bacterial counts in culture and infected wounds, respectively. In the present study, the cellulase was also highly efficient with ceftazidime and reduced the MBEC 32 to 128 times. Several studies found the ability of different enzymes to decrease the MBEC of antibiotics, including ceftazidime [36, 38, 43, 49]. The synergistic effect achieved by using enzyme/antibiotic combination can rapidly enhance anti-biofilm activity and help prevent or delay the emergence of resistance.
Our data showed the anti-biofilm activity of cellulase, separately and in combination with ceftazidime, against P. aeruginosa. So, further studies are recommended to perform on it. The cytotoxic effect of the enzyme must be evaluated. Given the polymicrobial nature of biofilms, it is proposed to investigate the cellulase’s efficacy on the destruction of mixed-species biofilms. It must be addressed whether the enzyme increases the effectiveness of all antibiotics or just certain classes. Moreover, the effects of the enzyme on immune system response to clear infections should be examined. Thus, these are just some of the questions needed to be answered before clinical applications of cellulase can be explored.
Conclusions
Considering the anti-biofilm properties of glycoside hydrolase cellulase in this study, including significant degradation of P. aeruginosa biofilm and a significant decrease in the ceftazidime MBEC, it appears to be a promising candidate, either as a single agent or as in combination with antibiotics for disturbing/dispersing biofilm and making highly resistant wound infections susceptible to traditional treatments.
Supplementary Information
Additional file 1. The effects of selected concentrations of cellulase (0.01-10 U/mL) against biofilm embedded P. aeruginosa isolates and ATCC PAO1 strain.
Acknowledgements
The authors would like to appreciate the staffs of hospitals and all colleagues at the Department of Microbiology, Golestan University of Medical Sciences, Gorgan, Iran, for their cooperation in the present study.
Abbreviations
- CAMHB
Cation-adjusted Mueller-Hinton broth
- CLSI
Clinical and Laboratory Standards Institute
- CV
Crystal violet
- EPSs
Extracellular polymeric substances
- GH
Glycoside hydrolase
- HI
Heat-inactivated
- MBC
Minimum bactericidal concentration
- MBEC
Minimum biofilm eradication concentration
- MHB
Mueller–Hinton broth
- MIC
Minimum inhibitory concentration
- NIH
National Institutes of Health
- TSB
Tryptic soy broth
Authors' contributions
AA and EK made substantial contributions to the study design, data analysis, interpretation of results, and the drafting of the paper. AA drafted the manuscript and critically revised it. AJ and AI helped in data entry, data analysis, and the drafting of the paper. The author(s) read and approved the final manuscript.
Funding
The present study was financially supported by Golestan University of Medical Sciences, Gorgan, Iran, with Grant Number 970211010.
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All stages of this study were approved by the Ethics Committee of Golestan University of Medical Sciences with the ethical code number IR.GOUMS.REC.1397.020. All methods were carried out under relevant guidelines and regulations. The experiments were performed on previously isolated bacteria from clinical samples of hospitalized patients. The patients did not directly participate in this project. Ethics Committee of Golestan University of Medical Sciences with the ethical code number IR.GOUMS.REC.1397.020 waived the need for informed consent from the patients whose clinical samples were used for this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Esmat Kamali, Email: parya.kamali71@gmail.com.
Ailar Jamali, Email: jamali@goums.ac.ir.
Ahdieh Izanloo, Email: ahdieh.izanloo76@gmail.com.
Abdollah Ardebili, Email: ardebili_abdollah57@yahoo.com.
References
- 1.Fleming D, Chahin L, Rumbaugh K. Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob Agents Chemother. 2017;61(2). [DOI] [PMC free article] [PubMed]
- 2.Muhsin J, Ufaq T, Tahir H, Saadia A. Bacterial biofilm: its composition, formation and role in human infections. J Microbiol Biotechnol. 2015;4:1–14. [Google Scholar]
- 3.Ghannoum M, O’Toole G. Microbial Biofilms. Washington DC: American Society for Microbiology; 2004. [Google Scholar]
- 4.Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med. 2013;3(4):a010306. doi: 10.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-γ-mediated macrophage killing. J Immunol. 2005;175(11):7512–8. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- 6.Taylor PK, Yeung AT, Hancock RE. Antibiotic resistance in Pseudomonas aeruginosa biofilms: towards the development of novel anti-biofilm therapies. J Biotechnol. 2014;191:121–30. doi: 10.1016/j.jbiotec.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Singh S, Singh SK, Chowdhury I, Singh R. Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol J. 2017;11:53. doi: 10.2174/1874285801711010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamal M, Ahmad W, Andleeb S, Jalil F, Imran M, Nawaz MA, Hussaina T, Alid M, Rafiqa M, Atif Kamil M. Bacterial biofilm and associated infections. J Chinese Med Assoc. 2018;81(1):7–11. doi: 10.1016/j.jcma.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 9.Kamali E, Jamali A, Ardebili A, Ezadi F, Mohebbi A. Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa. BMC Res Notes. 2020;13(1):27. doi: 10.1186/s13104-020-4890-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tolker-Nielsen T. Pseudomonas aeruginosa biofilm infections: from molecular biofilm biology to new treatment possibilities. APMIS Suppl. 2014;122:1–51. doi: 10.1111/apm.12335. [DOI] [PubMed] [Google Scholar]
- 11.Pournajaf A, Razavi S, Irajian G, Ardebili A, Erfani Y, Solgi S, Yaghoubi S, Rasaeian A, Yahyapour Y, Kafshgari R, et al. Integron types, antimicrobial resistance genes, virulence gene profile, alginate production and biofilm formation in Iranian cystic fibrosis Pseudomonas aeruginosa isolates. Infez Med. 2018;26(3):226–36. [PubMed] [Google Scholar]
- 12.Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. 2013;67(3):159–73. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 13.Church D, Elsayed S, Reid O, Winston B, Lindsay R. Burn wound infections. Clin Microbiol Rev. 2006;19(2):403–34. doi: 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 2014;10(7):e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metcalf DG, Bowler PG. Biofilm delays wound healing: a review of the evidence. Burn Trauma. 2013;1(1):2321-3868.113329. doi: 10.4103/2321-3868.113329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nidadavolu P, Amor W, Tran PL, Dertien J, Colmer-Hamood JA, Hamood AN. Garlic ointment inhibits biofilm formation by bacterial pathogens from burn wounds. J Med Microbiol. 2012;61(5):662–71. doi: 10.1099/jmm.0.038638-0. [DOI] [PubMed] [Google Scholar]
- 17.Nagraj AK, Gokhale D. Bacterial biofilm degradation using extracellular enzymes produced by Penicillium janthinellum EU2D-21 under submerged fermentation. Adv Microbiol. 2018;8(9):687–98. [Google Scholar]
- 18.Naumoff D. Hierarchical classification of glycoside hydrolases. Biochem (Mosc) 2011;76(6):622–35. doi: 10.1134/S0006297911060022. [DOI] [PubMed] [Google Scholar]
- 19.Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73(2):310–47. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–33. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 21.Rajasekharan SK, Ramesh S. Cellulase inhibits Burkholderia cepacia biofilms on diverse prosthetic materials. Pol J Microbiol. 2013;62(3):327–00. [PubMed] [Google Scholar]
- 22.Loiselle M, Anderson KW. The use of cellulase in inhibiting biofilm formation from organisms commonly found on medical implants. Biofouling. 2003;19(2):77–85. doi: 10.1080/0892701021000030142. [DOI] [PubMed] [Google Scholar]
- 23.Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing, Wayne P., 28th ed; 2019,M100.
- 24.Overhage J, Campisano A, Bains M, Torfs EC, Rehm BH, Hancock RE. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect Immun. 2008;76(9):4176–82. doi: 10.1128/IAI.00318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naparstek L, Carmeli Y, Navon-Venezia S, Banin E. Biofilm formation and susceptibility to gentamicin and colistin of extremely drug-resistant KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2014;69(4):1027–34. doi: 10.1093/jac/dkt487. [DOI] [PubMed] [Google Scholar]
- 26.Tetz GV, Artemenko NK, Tetz VV. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob Agents Chemother. 2009;53(3):1204–9. doi: 10.1128/AAC.00471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins M, Henriques M, Lopez-Ribot JL, Oliveira R. Addition of DNase improves the in vitro activity of antifungal drugs against Candida albicans biofilms. Mycoses. 2012;55(1):80–5. doi: 10.1111/j.1439-0507.2011.02047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167–93. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–22. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 30.Gawande PV, Leung KP, Madhyastha S. Antibiofilm and antimicrobial efficacy of DispersinB®-KSL-W peptide-based wound gel against chronic wound infection associated bacteria. Curr Microbiol. 2014;68(5):635–41. doi: 10.1007/s00284-014-0519-6. [DOI] [PubMed] [Google Scholar]
- 31.Püllen R, Popp R, Volkers P, Füsgen I. Prospective randomized double-blind study of the wound‐debriding effects of collagenase and fibrinolysin/deoxyribonuclease in pressure ulcers. Age Ageing. 2002;31(2):126–30. doi: 10.1093/ageing/31.2.126. [DOI] [PubMed] [Google Scholar]
- 32.Nemoto K, Hirota K, Murakami K, Taniguti K, Murata H, Viducic D, Miyake Y. Effect of Varidase (streptodornase) on biofilm formed by Pseudomonas aeruginosa. Chemotherapy. 2003;49(3):121–5. doi: 10.1159/000070617. [DOI] [PubMed] [Google Scholar]
- 33.Ramundo J, Gray M. Enzymatic wound debridement. J Wound Ostomy Continence Nurs. 2008;35(3):273–80. doi: 10.1097/01.WON.0000319125.21854.78. [DOI] [PubMed] [Google Scholar]
- 34.Trizna E, Bogachev MI, Kayumov A. Degrading of the Pseudomonas aeruginosa biofilm by extracellular levanase SacC from Bacillus subtilis. BioNanoScience. 2019;9(1):48–52. [Google Scholar]
- 35.Baker P, Hill PJ, Snarr BD, Alnabelseya N, Pestrak MJ, Lee MJ, Jennings LK, Tam J, Melnyk RA, Parsek MR, et al. Exopolysaccharide biosynthetic Melnyk glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci Adv. 2016;2(5):e1501632. doi: 10.1126/sciadv.1501632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banar M, Emaneini M, Satarzadeh M, Abdellahi N, Beigverdi R, van Leeuwen WB, Jabalameli F. Evaluation of mannosidase and trypsin enzymes effects on biofilm production of Pseudomonas aeruginosa isolated from burn wound infections. PloS one. 2016;11(10):e0164622. doi: 10.1371/journal.pone.0164622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daboor SM, Raudonis R, Cohen A, Rohde JR, Cheng Z. Marine bacteria, a source for alginolytic enzyme to disrupt Pseudomonas aeruginosa biofilms. Marine Drugs. 2019;17(5):307. doi: 10.3390/md17050307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banar M, Emaneini M, Beigverdi R, Pirlar RF, Farahani NN, van Leeuwen WB, van Leeuwen WB, Jabalameli F. The efficacy of lyticase and β-glucosidase enzymes on biofilm degradation of Pseudomonas aeruginosa strains with different gene profiles. BMC Microbiol. 2019;19(1):291. doi: 10.1186/s12866-019-1662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alhede M, Bjarnsholt T, Givskov M, Alhede M. Pseudomonas aeruginosa biofilms: mechanisms of immune evasion. In: Adv Appl Microbiol. Elsevier; 2014. p. 1-40. [DOI] [PubMed]
- 40.Fleming D, Rumbaugh K. The consequences of biofilm dispersal on the host. Sci. Rep. 2018;8(1):1–7. doi: 10.1038/s41598-018-29121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fleming D, Redman W, Welch GS, Mdluli NV, Rouchon CN, Frank KL, Rumbaugh KP. Utilizing glycoside hydrolases to improve the quantitation and visualization of biofilm bacteria. Biofilm. 2020;2:100037. doi: 10.1016/j.bioflm.2020.100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013;121:1–58. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 43.Dosler S, Karaaslan E. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides. 2014;62:32–7. doi: 10.1016/j.peptides.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 44.Toté K, Berghe DV, Deschacht M, De Wit K, Maes L, Cos P. Inhibitory efficacy of various antibiotics on matrix and viable mass of Staphylococcus aureus and Pseudomonas aeruginosa biofilms. Int J Antimicrob Agents. 2009;33(6):525–31. doi: 10.1016/j.ijantimicag.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Hengzhuang W, Ciofu O, Yang L, Wu H, Song Z, Oliver A, Høiby N. High β-lactamase levels change the pharmacodynamics of β-lactam antibiotics in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2013;57(1):196–204. doi: 10.1128/AAC.01393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cordeiro AL, Hippius C, Werner C. Immobilized enzymes affect biofilm formation. Biotechnol Lett. 2011;33(9):1897–904. doi: 10.1007/s10529-011-0643-3. [DOI] [PubMed] [Google Scholar]
- 47.Otani S, Hiramatsu K, Hashinaga K, Komiya K, Umeki K, Kishi K, Kadota JI. Sub-minimum inhibitory concentrations of ceftazidime inhibit Pseudomonas aeruginosa biofilm formation. J Infect Chemother. 2018;24(6):428–33. doi: 10.1016/j.jiac.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 48.Pestrak MJ, Baker P, Dellos-Nolan S, Hill PJ, da Silva DP, Silver H, Lacdao I, Raju D, Parsek MR, Wozniak DJ, et al. treatment with the Pseudomonas aeruginosa glycoside hydrolase PslG combats wound infection by improving antibiotic efficacy and host innate immune activity. Antimicrob Agents Chemother. 2019;63(6). [DOI] [PMC free article] [PubMed]
- 49.Fanaei Pirlar R, Emaneini M, Beigverdi R, Banar M, van Leeuwen B, Jabalameli W. Combinatorial effects of antibiotics and enzymes against dual-species Staphylococcus aureus and Pseudomonas aeruginosa biofilms in the wound-like medium. Plos one. 2020;15(6):e0235093. doi: 10.1371/journal.pone.0235093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The effects of selected concentrations of cellulase (0.01-10 U/mL) against biofilm embedded P. aeruginosa isolates and ATCC PAO1 strain.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.