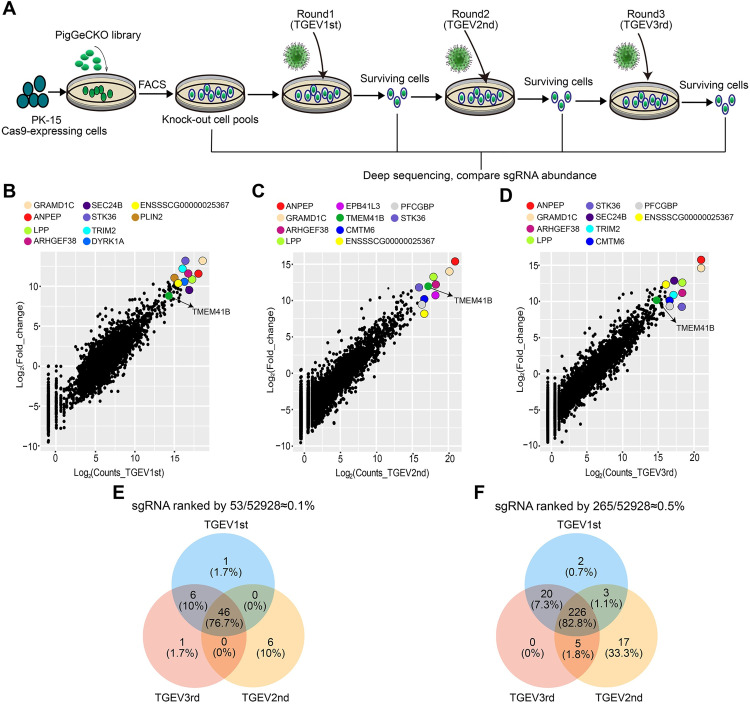
Fig 1. Genome-wide CRISPR screening to identify genes associated with alpha-CoVs TGEV-induced cell death.
(A) Identification of TGEV host factors using the porcine genome-scale CRISPR/Cas9 knockout (PigGeCKO) library. Transformed PK-15-Cas9 cells were either mock-treated or challenged with TGEV (MOI = 0.001). Surviving cells from each round of virus challenge were isolated, with the subsequent PCR amplification and sequencing of sgRNA. (B-D) Scatter plots showing sgRNA-targeted sequence frequencies and the extent of enrichment in transformed PK-15-Cas9 cells (mock-treated versus TGEV infected) in three rounds of TGEV screening; (B) first, (C) second and (D) third, respectively. Counts_TGEV1st/Counts_TGEV2nd and Counts_TGEV3rd are the average reads from the paired-end sequencing for each round. Log2(fold change) is the median log 2 ratio between normalized sgRNA count of TGEV challenged and mock-treated populations. (E and F) Venn diagrams indicate the scope of overlapping enrichment in specific sgRNA targeting sequences for the three TGEV screening rounds amongst the top (E) ~0.1% and (F) ~0.5% of averaged sgRNAs reads. FACS, Fluorescence Activated Cell Sorting; TGEV, Transmissible gastroenteritis virus; sgRNA, small guide RNA.