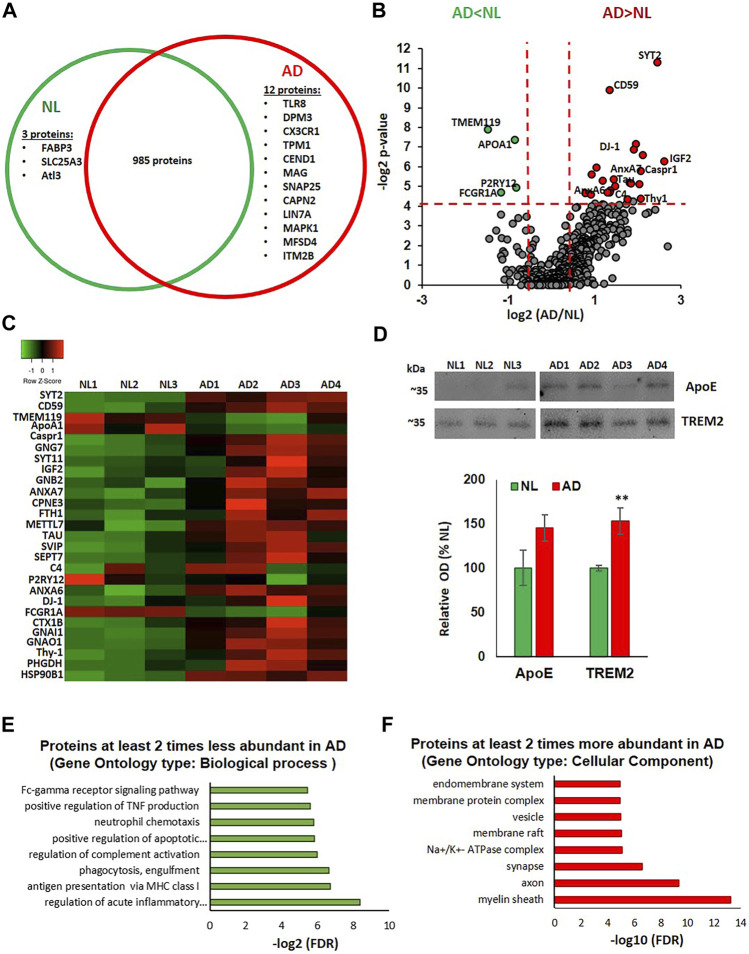
FIGURE 2.
Proteomic analysis of microglial EVs from normal/low pathology and late-stage AD cases. (A) Venn diagram representing microglial proteins differentially detected in NL and AD groups. (B) Volcano plot showing a degree of differential expression of proteins in NL and AD groups. Vertical red dotted lines separate proteins, which are two or more times less abundant in AD (far left segment) and two or more times more abundant in AD (far right segment). Horizontal red dotted line separates the top part of the plot containing dots representing proteins whose abundances are significantly different between NL and AD groups (p < 0.05) and the bottom part (no significant changes). Proteins that are significantly different between AD and NL groups are color-coded: two or more times more abundant in AD are presented in red, two or more times less abundant in green. (C) Heat map of proteins significantly up- and downregulated in AD compared to the NL group across seven evaluated human cases. (D) Immunoblotting of CD11b-IP samples with antibodies against ApoE and TREM2. Representative images are in the upper section and the composite data are in the lower section (*p < 0.05). (E) Gene ontology enrichment bioinformatic analysis of proteins two or more times less abundant in AD compared to NL. (F) Gene ontology enrichment bioinformatic analysis of proteins two or more times more abundant in AD compared to NL.