Abstract
Present study utilised textile soil isolated bacterium Pseudomonas stutzeri to synthesise extracellular silver nanoparticles (AgNPs) under optimised conditions. The synthesised AgNPs were characterised using ultraviolet‐visible spectroscopy, Fourier transform infrared spectroscopy (FTIR) and transmission electron microscopy (TEM). Optimisation showed AgNPs synthesis within 8 h using 2mM Ag nitrate at pH9, temperature 80°C and maximum absorbance toward 400 nm. TEM analysis revealed spherical shape AgNPs and reduction in size upto 8 nm was observed under optimised conditions. FTIR spectra confirmed presence of proteins bound to AgNPs act as reducing agent. AgNPs showed strong antibacterial activity against multi‐drug resistant (MDR) Escherichia coli and Klebsiella pneumoniae as demonstrated by disc diffusion and colony forming unit assays. Zone of inhibition increased with increasing concentration of AgNPs with maximum of 19 mm against E. coli and 17 mm against K. pneumoniae at concentration of 2 μg/disc. Furthermore, AgNPs did not show any cytotoxic effects on human epithelial cells as demonstrated by 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl tetrazolium bromide assay even at 2 μg/ml concentration of AgNPs. The results of the present study suggest that AgNPs can be synthesised rapidly under optimised conditions and show strong antimicrobial property against MDR pathogens without having toxicity effect on human epithelial cells.
Inspec keywords: ultraviolet spectra, proteins, transmission electron microscopy, infrared spectra, Fourier transform spectra, visible spectra, microorganisms, toxicology, cellular biophysics, biomedical materials, antibacterial activity, nanomedicine, nanofabrication, nanoparticles, silver
Other keywords: 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl tetrazolium bromide assay; human epithelial cells; cytotoxic effects; K. pneumoniae; colony forming unit counting assays; disc diffusion; Klebsiella pneumoniae; Escherichia coli; multidrug resistant; stabilising agent; reducing agent; proteins; parametric optimisation; TEM; transmission electron microscopy; FTIR spectra; Fourier transform infrared spectroscopy; ultraviolet‐visible spectroscopy; bacterium; cytotoxicity properties; antimicrobial properties; textile soil; Pseudomonas stutzeri; silver nanoparticle synthesis
1 Introduction
Nanotechnology, deals with the synthesis and applications of nanoscale materials in diverse interdisciplinary fields such as physics, chemistry, biology, medicine and agriculture [1]. Numerous metals such as silver (Ag) [2], gold [3], copper [4], zinc [5], iron [6], palladium [7], selenium [8] and titanium [9] have been investigated at the nanoscale. Among all the metallic nanoparticles Ag nanoparticles (AgNPs) have received greatest attention because of strong antimicrobial action [10] and hence used in nanomedicine.
Vast applications and rapid utilisation of engineered nanoparticles would inevitably lead to the release of these materials into the environment and cause toxicity to different organisms. Various techniques such as physical and biological means have been utilised for the synthesis of AgNPs. However, biological route of synthesis using bacteria, yeast, fungi, algae and plants is gaining attention due to cost‐effectiveness, least toxicity and better optimisation control [11].
Microorganisms utilise both extracellular and intracellular routes for the synthesis of nanoparticles [12]. Extracellular biosynthesis is preferable as it reduces the complex downstream processing than the intracellular biosynthesis which requires additional steps such as cell lysis by ultra‐sonicator or suitable detergent treatment for purification of synthesised nanoparticles [13]. A number of bacterial species including Escherichia coli [14], Ureibacillus thermosphaericus [15], Staphylococcus aureus [16], Bacillus thuringiensis [17], Pseudomonas aeruginosa [18] and Bacillus cereus [19] have been explored to synthesise AgNPs. The limitation behind bacteriogenic synthesis of nanoparticle is slow rate of synthesis, non‐uniform size of nanoparticles and polydispersity [20]. Moreover, optimisation studies of various parameters such as Ag nitrate (AgNO3) concentration, pH and temperature for enhanced and rapid synthesis of AgNPs are lacking.
The growing resistance of pathogenic bacterial strains to traditional antibacterial treatments has encouraged the development of alternate strategies to control infections. The development of non‐toxic methods of synthesising nanoparticles would be a major step in nanotechnology to allow their application in medicine. Being strong antibacterial, AgNPs because of its small size with increased surface have been used as biocide in different formulations as well as in increasing number of consumer and medical products [21]. AgNPs were reported to have antibacterial activity against various pathogenic bacteria both Gram positive and Gram negative [22]. Several mechanisms including penetration of bacterial cell wall, damaging the cell membrane, generation of free radicals, inhibition of signal transduction pathways and DNA damage have been reported [23, 24].
Nanomaterials synthesised have shown some profound effect on human health. Nanoparticles have ability to enter human body in various ways and able to cause toxicity mainly by generation of reactive oxygen species and genotoxicity [25]. Many nanoparticles have also been reported to induce inflammatory response through activation of expression of tumour necrosis factor alfa. Nanoparticles can also lead to an increased expression of other pro‐inflammatory cytokines such as interleukins 2, 6, 8 and 10 (IL‐2, IL‐6, IL‐8 and IL‐10) [26, 27].
The present study deals with the extracellular biosynthesis of AgNPs, using cell filtrate of P. stutzeri isolated from soil. Furthermore, characterisation and optimisation for rapid AgNPs synthesis were carried out. Antimicrobial and toxicity studies have also been elucidated.
2 Materials and methods
2.1 Chemical, culture media and bacterial strains
The chemical and culture media used in the study were purchased from Sisco Research Laboratories (SRL), India and Himedia, India, respectively. The test organisms including E. coli and Klebsiella pneumonia were procured from All India Institute of Medical Science (AIIMS), New Delhi.
2.2 Sample collection and isolation of bacteria
The soil sample was collected from textile area near Sanganer, Jaipur, Rajasthan, India at 10 cm depth in sterilised zip‐lock bag. The collected soil sample was stored in ice box and then transported to the laboratory within 6 h. About 1 g of soil was serially diluted and spread over nutrient agar followed by incubation at 37°C for overnight. Ten colonies were picked randomly and sub‐cultured to obtain pure culture.
2.3 Screening for extracellular synthesis of AgNPs
The ten isolates were freshly inoculated in 100 ml nutrient broth in Erlenmeyer flask and incubated in orbital shaker for 24 h at 37°C and at 200 rpm. Culture filtrates were obtained by centrifugation at 10,000 rpm for 10 min. The supernatant was challenged with 2.0 mM AgNO3 solution and incubated in dark condition at 37°C. The isolate showing maximum AgNPs synthesis was characterised by morphological and biochemical tests as per the procedures outlined in Bergey's Manual of Determinative Bacteriology [28]. The bacterium was also characterised based on 16S rDNA technique at Xcleries genomics laboratory. The bacterium was identified as P. stutzeri.
2.4 Optimisation studies for rapid AgNPs synthesis
Different reaction parameters such as AgNO3 concentration, pH and temperature were optimised to obtain maximum production of AgNPs [29].
2.4.1 Effect of AgNO3 concentration
Nanoparticles synthesis is dependent on substrate concentration. AgNPs were synthesised using different concentrations of AgNO3 from 0.2 to 2.5 mM. The sample was analysed with ultraviolet–visible (UV–vis) absorption spectroscopy at regular time interval from 24 to 144 h.
2.4.2 Effect of pH
pH plays a very important role in growth and enzyme production which is required for the biosynthesis of AgNPs. Different pH values ranging from 3.0 to 11.0 were used to study the influence of pH on AgNPs production and UV–vis absorption spectra were recorded from 24 to 144 h after addition of AgNO3.
2.4.3 Effect of temperature
Optimisation studies with respect to temperature were carried out with temperature ranging from 20 to 80°C for AgNPs production. UV–vis absorption spectra were taken at different time interval from 0 to 24 h.
2.5 Characterisation of AgNPs
2.5.1 UV–vis spectroscopy analysis
AgNO3 treated culture filtrate of P. stutzeri was monitored for reduction of Ag ions by reading the sample aliquots (1 ml) at different time intervals on UV‐vis spectrophotometer (Thermo scientific/Biomate 3S) at a resolution of 1 nm.
2.5.2 FTIR spectroscopy analysis
The interaction between protein and AgNPs was analysed by Fourier transform infrared (FTIR) spectroscopy. The bio‐transformed products present in culture supernatant were freeze‐dried and FTIR spectrum was recorded on FTIR (Shimazdu, India) in the range of 400–4000 cm−1 at a resolution of 4 cm−1.
2.5.3 Transmission electron microscopy (TEM) analysis
Morphological characterisation including shape and size of AgNPs synthesised by P. stutzeri culture supernatant was done by taking a drop of solution containing AgNPs loaded on copper grids in transmission electron microscope (Jeol, USA). TEM micrographs were taken and size and shapes were analysed.
2.6 Determination of antibacterial activity
2.6.1 Antibacterial activity by disc diffusion assay
Antibacterial activity of AgNPs synthesised from P. stutzeri was performed by disc diffusion method against E. coli and K. pneumoniae. In brief, overnight grown bacterial cultures were spread onto Muller‐Hinton agar followed by 10 µl of AgNPs (from 50, 100 and 200 µg/ml concentration of AgNPs) soaked sterile paper discs were kept on nutrient agar so as to achieve different 0.5, 1 and 2 µg of AgNPs/disc. The plates were kept overnight at 37°C and zone of inhibition was measured. Ampicillin (30 µg/disc) was used to show E. coli and K. pneumoniae as multi‐drug resistant (MDR) strains [30].
2.6.2 Antibacterial activity in suspension
About 105 colony forming units (CFUs) per millilitre of E. coli and K. pneumoniae in phosphate buffer saline (PBS) and different concentrations of AgNPs (0.5, 1, 1.5 and 2 µg/ml) were mixed and incubated at 37°C for 1 h. The samples were then plated on nutrient agar and incubated at 37°C. The numbers of the viable cells were determined by colony counting and 1 ml of pure PBS was used as a negative control. Bacterial killing percentage (% kill) was defined as (N PBS −N sample)/N PBS × 100, where N PBS and N sample represent the number of bacteria in PBS and the sample, respectively [31].
2.7 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl tetrazolium bromide (MTT) viability assay
Cell viability was assessed by MTT assays. For MTT assay, 2 × 103 cells/well were seeded into sterile 96‐well plate for 24 h and then exposed to increasing concentrations of AgNPs (0.5, 1, 1.5 and and 2 µg/ml). After 24 h of exposure, cells were washed with PBS and 0.5 μg/μl MTT (Sigma) was added and cells incubated for additional 4 h at 37°C in the dark. Medium was removed and formazan crystals were dissolved in Dimethyl Sulphoxide (DMSO) and cellular metabolism was determined by monitoring the colour development at 570 nm, in a multi‐well scanning spectrophotometer. Cells without AgNPs exposure were used as control. Cells viability percentage (%) was calculated as (sample Abs570–655 nm)/(control Abs570–655 nm) × 100 [32].
2.8 Hemolysis assay
Hemolytic activity of AgNPs was evaluated by measuring the hemoglobin release from the erythrocytes [red blood cell (RBC)] on interaction with nanoparticles under in vitro conditions. About 5 ml blood was collected from human volunteer in a tube containing ethylenediaminetetraacetic acid and was centrifuged at 2000 rpm for 10 min. The supernatant and buffy coat was discarded and the pellet containing RBCs were washed with PBS. About 108 cells of RBCs diluted in PBS were added to different concentrations of AgNPs (50, 100 and 200 µg/ml) and incubated at 37°C with mild shaking for 1 h. Then, the tubes were centrifuged at 2000 rpm for 10 min and the supernatant were read at 540 nm. RBCs in 1% triton X‐100 were taken for complete lysis and considered as positive control. Percentage hemolysis (%) was calculated as (Abs540 nm in the AgNP solution−Abs540 nm in PBS)/(Abs540 nm in 1% Triton X‐100−Abs540 nm in PBS) × 100 [32].
3 Results and discussion
3.1 Molecular identification of bacterial isolate
Bacteria were isolated from textile soil samples of Jaipur, Rajasthan, India by serial dilution and sub‐culturing. Randomly, ten bacterial isolates were selected and screened for AgNPs synthesis. Strain showing AgNP formation by UV–vis spectrophotometer was characterised. Morphological and biochemical characteristics of isolate have been outlined in Table 1. The partially amplified (803 bp) and sequenced 16S rRNA gene was uploaded to the National Center for Biotechnology Information website to search for similarity to known DNA sequences and to confirm the species of the isolate. The nucleotide sequences coding for the 16S rRNA gene after Basic Local Alignment Search Tool query revealed that this gene is 99% homologous to P. stutzeri (BPRIST027). The nucleotide sequences coding for the 16S rRNA gene of P. stutzeri have been submitted to the GenBank database under accession numbers KU530223.
Table 1.
Morphological and biochemical characteristics of Pseudomonas stutzeri
| Characters | P. stutzeri |
|---|---|
| morphology | |
| shape | rod |
| gram stain | − |
| endospore | − |
| IMVIC | |
| indole | − |
| methyl red | − |
| Voges–Proskauer | + |
| citrate | + |
IMVIC, Indole Methyl red Voges‐Proskauer and Citrate test
3.2 Extracellular synthesis of AgNPs
Visual observation of AgNO3 showed a change in colour intensity from light (Fig. 1 B inset) to dark (Fig. 1 C inset). No change was observed in culture supernatant without AgNO3 (Fig. 1 B inset). The appearance of a dark colour (Fig. 1 C inset) in AgNO3 treated culture supernatant suggested the formation of AgNPs. The oscillation waves of electrons of AgNPs are in resonance with the light waves gives rise to a unique surface plasmon resonance (SPR) absorption band (400–440 nm), which is also the origin of the observed colour [33]. The extracellular synthesis of AgNPs using P. stutzeri was monitored in UV–vis spectroscopy. Graph shown a strong peak at 430 nm (Fig. 1), indicated an SPR, having nanoparticles with sizes ranging from 2 to 100 nm. Similar results were reported by Singh [30], who also synthesised AgNPs from Acinetobacter calcoaceticus. The precise mechanism involved in bacteria mediated synthesis of AgNPs has not been known. However, it can be supposed that nitrate reductase secreted by microbes help in the bioreduction of metal ions to metal nanoparticles [34]. Furthermore, Kalimuthu et al. [35] confirmed the Bacillus licheniformis mediated secretion of nitrate reductase responsible for the reduction of Ag+ to nanoparticles.
Fig. 1.
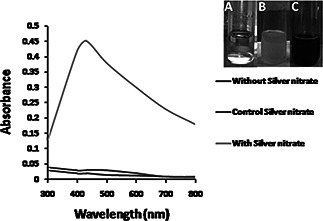
UV‐Vis spectra of AgNPs synthesized using culture supernatant of P. stutzeri at 2mM AgNO3, pH 7 and temperature 30o C.
a Blank AgNO3 solution
b Blank culture supernatant
c Dark color AgNPs solution
3.3 Optimisation studies for AgNPs production
3.3.1 Effect of AgNO3 concentration
Different concentrations of AgNO3 solution (0.2–2.5 mM) was used to synthesise AgNPs using P. stutzeri culture supernatant and monitored at regular interval from 24 to 144 h. At 0.2mM AgNO3 concentration synthesis of AgNPs is very low which increases up to 2 mM AgNO3 at 120 h of incubation (Fig. 2 a). Amount of AgNPs decreased after 144 h of incubation indicating complete reduction of Ag+ at 120 h. Maximum peak at 2 mM concentration is about 430 nm (Fig. 2 b) and further increase in AgNO3 concentration led to decrease in the synthesis of AgNPs. This can be explained on the basis of enzyme–substrate kinetics where active site of enzyme catalysing reduction is saturated with the Ag ions and no more site is available for ions to be reduced; therefore, no further increase in AgNPs synthesis at higher concentration of AgNO3 [36].
Fig. 2.
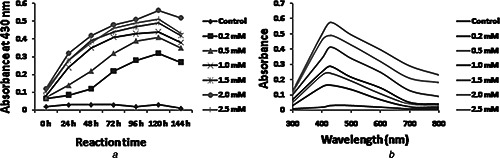
Optimisation of AgNO3 concentration for AgNP synthesis
a Time‐dependent AgNP synthesis at 30°C using different concentrations of AgNO3
b UV–vis spectra of AgNP synthesis at 30°C after 120 h obtained with different concentrations of AgNO3
3.3.2 Effect of pH
To optimise the pH for maximum AgNPs synthesis, AgNO3 was added in the culture supernatant having different pH (3, 5, 7, 9 and 11). AgNPs were found to be synthesised within 48 h at pH 9 and remain stable up to 144 h (Fig. 3 a). UV–vis spectra demonstrated maximum absorbance peak of 430 nm (Fig. 3 b). It may be due to increased availability of OH− at pH 9, which helps in complete reduction of Ag+ into AgNPs by providing electrons [36]. However, increase in pH up to 11 and decreased synthesis of AgNPs were observed due to inactivation of enzyme responsible for AgNPs synthesis. Our data are in agreement with Sintubin et al. [37], who also reported enhanced AgNPs production at alkaline pH using lactic acid bacteria due to more competition between H+ and Ag+ for negatively charged OH−. On the contrary, acidic pH led to increased H+ concentration which may denature enzymes responsible for AgNPs synthesis [38].
Fig. 3.
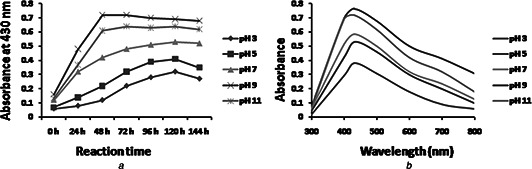
Optimisation of pH for AgNP synthesis
a Time‐dependent AgNP synthesis at 30°C using 2 mM concentrations of AgNO3 at different pH
b UV–vis spectra of AgNPs synthesis at 30°C after 120 h at different pH
3.3.3 Effect of temperature
Temperature is an important factor affecting AgNPs production. Effect of temperatures on AgNPs production by P. stutzeri was carried out at different temperatures from 20 to 80°C with a difference of 20°C, 2 mM AgNO3, pH 9 and monitored at regular time interval. At 80°C, synthesis of AgNPs was observed within 8–10 h and remains stable for longer time period indicated rapid and stabilised synthesis (Fig. 4 a). There was little shift in peak from 430 nm toward 400 nm as temperature increases from 40 to 80°C (Fig. 4 b). This may be due to increase in kinetic energy of the AgNPs in the solution at high temperature, which leads to faster synthesis rate [39]. Similar results have been found by Gurunathan et al. [40], who used E. coli for AgNPs synthesis and found maximum synthesis at 5 mM AgNO3, 60°C temperature and pH 10.
Fig. 4.
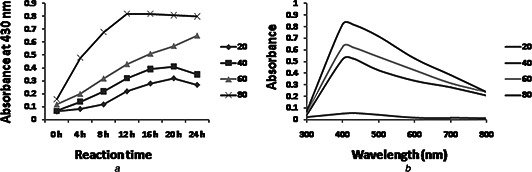
Optimisation of temperature for AgNP synthesis
a Time‐dependent AgNP synthesis at different temperatures using 2 mM concentrations of AgNO3 at pH 9
b UV–vis spectra of AgNPs synthesis at different temperatures after 120 h at pH 9
3.4 TEM characterisation of synthesised AgNPs
To determine the shape, size and morphology of P. stutzeri mediated synthesised AgNPs under optimised conditions, TEM measurements were carried out. TEM micrograph revealed that the nanoparticles are spherical in shape and well dispersed without agglomeration. Selected area electron diffraction (SAED) pattern revealed the crystalline nature of AgNPs synthesised using culture supernatant of P. stutzeri (Fig. 5 b inset and 5d inset). The average particle size of AgNPs synthesised by P. stutzeri ranges from 15 to 20 nm (Figs. 5 A and B). However, under optimised conditions, smaller size of AgNPs was synthesised with average size of 8 nm (Figs. 5 C and D). This reduction of AgNPs size will results in increased surface area of contact with microorganisms and make it better bactericidal [41]. In another study, AgNPs of average size 27.5 nm were synthesised using culture filtrate of P. aeruginosa [18]. Furthermore, the presence of capping material around each particle as revealed by well separated AgNPs will improve stability and increase chances of attachment on bacterial surface [42].
Fig. 5.
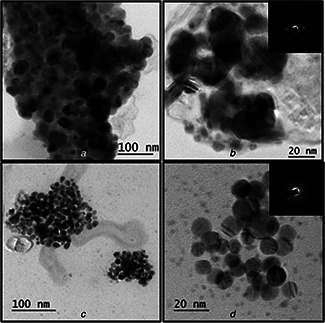
Morphology of AgNPs synthesised under normal and optimised conditions. TEM images for AgNPs synthesised at 30°C, pH 7 with 2 mM AgNO3 at low (A) and high magnification (B). TEM images for AgNPs synthesised at 80°C, pH 9 with 2 mM AgNO3 at low (C) and high magnification (D). Inset shows SAED pattern of AgNPs
3.5 FTIR characterisation of synthesised AgNPs
FTIR measurement of the dried and powdered sample was carried out to identify the presence of proteins surrounding AgNPs that could be responsible for synthesis and stabilisation of AgNPs. FTIR spectrum revealed the presence of peak around 1630 cm−1 which is assigned as absorption peak of –C=O and –C=C– stretching, and stretch vibration of –C=C– and is assigned to the amide I bonds of proteins (Fig. 6). The presence of peak at 1452 cm−1 may be assigned to symmetric stretching vibrations of –COO– groups of amino acid residues with free carboxylate groups in the protein. The peaks seen at 1380 and 1034 cm−1 correspond to –C–N stretching vibrations. The presence of band at 1538 and 1239 cm−1 assigned to binding vibrations of secondary amine and aliphatic amines, respectively. Hence, FTIR analysis confirmed the occurrence of capping protein around the AgNPs synthesised from culture supernatant of P. stutzeri. The presence of protein cap in nanoparticles help in stabilisation and binding to cell surface receptor results in increased binding and uptake of drug or genetic material on human cells [43, 44].
Fig. 6.
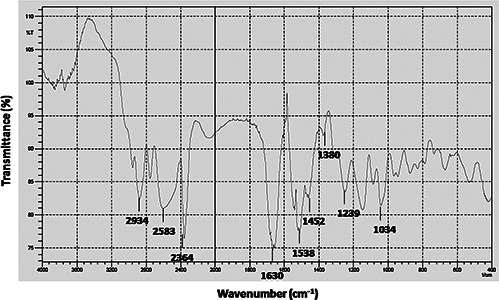
FTIR spectra of synthesised AgNPs
3.6 Antibacterial Assay of AgNPs against MDR E. coli and K. pneumoniae
Antibacterial assay of biosynthesised AgNPs was studied against MDR pathogenic strains (clinical isolates obtained from AIIMS, New Delhi) of E. coli and K. pneumoniae using disc diffusion method and zone of inhibition was depicted in Fig. 7. The results revealed that AgNPs were most effective against MDR E. coli. It was 19 mm at 2 µg/disc. For MDR K. pneumoniae, however, AgNPs showed a mild growth inhibitory effect of 17 mm even at 2 µg/disc when compared with MDR E. coli strain (Table 2). MDR E. coli and K. pneumoniae did not show zone of inhibition against ampicillin used as control (Fig. 7 a).
Fig. 7.
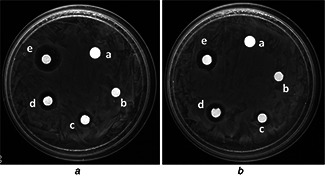
Zone of inhibition of AgNPs at (c) 0.5 μg, (d) 1 μg and (e) and 2 μg (e) against (A) E. coli and (B) K. pneumoniae. (a) Ampicillin was used as positive control for MDR E. coli and K. pneumoniae. (b) Water was used as negative control
Table 2.
Zone of inhibition of AgNPs produced by the Pseudomonas sp. against MDR E. coli and K. pneumoniae
| Sl. No | MDR bacterial strains | Zone of inhibition, mm ± scanning electron microscopy (SEM) | ||
|---|---|---|---|---|
| 0.5 µg/disc | 1 µg/ml | 2 µg/disc | ||
| 1. | E. coli | 11 ± 0.4 | 16 ± 0.8 | 19 ± 1.1 |
| 2. | K. pneumoniae | 7 ± 0.5 | 11 ± 0.6 | 17 ± 0.9 |
Furthermore, antibacterial activity of AgNPs in suspension was evaluated by counting CFUs of E. coli and K. pneumoniae after 1 h exposure to AgNPs (0.5–2 µg/ml). Fig. 8 shows that increasing concentration of AgNPs significantly augments the kill percentage against E. coli and K. pneumoniae. Ag+ from AgNPs can anchor to the negatively charged bacterial cell wall lead to perforation and results in cell lysis. Moreover, role of free radicals produced by AgNPs in contact with bacteria was also reported as demonstrated by electron spin resonance spectroscopy [10].
Fig. 8.
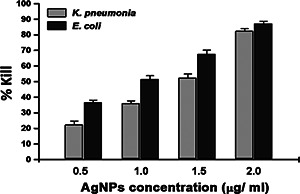
Kill percentages of AgNPs
3.7 In vitro cytotoxicity of AgNPs
To explore the cytotoxicity of AgNPs, the MTT assay was employed measuring the mitochondrial activity of HeLa cells. Cells were exposed to increasing concentrations of AgNPs (0.5–2 µg/ml) for 24 h. The MTT results of AgNPs did not show concentration‐dependent effects on the mitochondrial activity of HeLa cells (Fig. 9), indicating that the cells were unaffected by AgNPs synthesised using P. stutzeri at any of the employed concentrations. The selective toxicity of AgNPs toward bacterial cells as compared with HeLa cells can be explained in terms of the presence of transporter systems that may be responsible for differential binding and uptake of protein‐capped AgNPs conferring cytotoxicity [47].
Fig. 9.
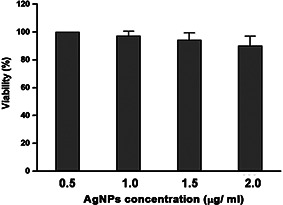
Concentration‐dependent cytotoxicity of AgNPs
3.8 In vitro hemolytic of AgNPs
Hemolytic activity of AgNPs was performed to evaluate the biocompatibility of the AgNPs on normal cells, especially RBCs. The hemolytic assay was performed using different concentrations of AgNPs (0.5–2 µg/ml) on RBCs. The absorbance values of RBCs in PBS and in 1% triton X‐100 was used along with the values of RBCs treated with AgNPs on the formula. It was found that AgNPs exhibit little hemolytic activity against RBCs at 2 µg/ml concentrations (Table 3). Our data are in correlation with the previous study done by Balaji et al. who reported that AgNPs (125 µg/ml) synthesised using aqueous extract of Rosa damascena petals did not show hemolytic activity against RBCs [48].
Table 3.
Hemolytic activity of AgNPs on RBCs
| Sl. No | Treatment | Percentage hemolysis ± SEM |
|---|---|---|
| 1. | control RBCs | 2.3 ± 0.03 |
| 2. | RBC+1% triton × 100 | 100 ± 0 |
| 3. | 0.5 µg/ml | 3.58 ± 0.1 |
| 4. | 1 µg/ml | 4.7 ± 0.08 |
| 5. | 2 µg/ml | 8.2 ± 0.11 |
4 Conclusion
This is the first report on biological synthesis of AgNPs using P. stutzeri, isolated from textile soil. The optimisation of reaction parameters resulted in monodispersed spherical AgNPs of size 8–10 nm. TEM was used to study the morphology, shape and size of biosynthesised nanoparticles. FTIR revealed the presence of protein cap on biosynthesised AgNPs. AgNPs exhibited excellent antimicrobial property on MDR E. coli and K. pneumonia. This eco‐friendly and cost‐effective extracellular biosynthesis of naturally protein‐capped AgNPs with potent antimicrobial activities from P. stutzeri has the potential to be utilised on a large scale for medical application.
5 Acknowledgment
We are indebted to the Amity University Rajasthan for providing necessary facilities and valuable support throughout the work.
6 References
- 1. Qu X. Alvarez P.J. Li Q.: ‘Applications of nanotechnology in water and wastewater treatment’, Water Res., 2013, 47, pp. 3931 –3946 (doi: 10.1016/j.watres.2012.09.058) [DOI] [PubMed] [Google Scholar]
- 2. Wei L. Lu J. Xu H. et al.: ‘Silver nanoparticles: synthesis, properties, and therapeutic applications’, Drug Discov. Today, 2015, 20, pp. 595 –601 (doi: 10.1016/j.drudis.2014.11.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Majdalawieh A. Kanan M.C. El‐Kadri O. et al.: ‘Recent advances in gold and silver nanoparticles: synthesis and applications’, J. Nanosci. Nanotechnol., 2014, 14, pp. 4757 –4780 (doi: 10.1166/jnn.2014.9526) [DOI] [PubMed] [Google Scholar]
- 4. Benavente E. Lozano H. Gonzalez G.: ‘Fabrication of copper nanoparticles: advances in synthesis, morphology control, and chemical stability’, Recent Pat. Nanotechnol., 2013, 7, pp. 108 –132 (doi: 10.2174/1872210511307020002) [DOI] [PubMed] [Google Scholar]
- 5. Shi L.E. Li Z.H. Zheng W. et al.: ‘Synthesis, antibacterial activity, antibacterial mechanism and food applications of ZnO nanoparticles: a review’, Food Additives Contaminants, A, 2014, 31, pp. 173 –186 (doi: 10.1080/19440049.2013.865147) [DOI] [PubMed] [Google Scholar]
- 6. Lee N. Hyeon T.: ‘Designed synthesis of uniformly sized iron oxide nanoparticles for efficient magnetic resonance imaging contrast agents’, Chem. Soc. Rev., 2012, 41, pp. 2575 –2589 (doi: 10.1039/C1CS15248C) [DOI] [PubMed] [Google Scholar]
- 7. Khazaei A. Rahmati S. Hekmatian Z. et al.: ‘A green approach for the synthesis of palladium nanoparticles supported on pectin: application as a catalyst for solvent‐free Mizoroki–Heck reaction’, J. Mol. Catal. A, Chem., 2013, 372, pp. 160 –166 (doi: 10.1016/j.molcata.2013.02.023) [DOI] [Google Scholar]
- 8. Husen A. Siddiqi K.S.: ‘Plants and microbes assisted selenium nanoparticles: characterization and application’, J. Nanobiotechnol., 2014, 12, pp. 1 –10 (doi: 10.1186/1477-3155-12-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rajakumar G. Rahuman A.A. Roopan S.M. et al.: ‘Fungus‐mediated biosynthesis and characterization of TiO2 nanoparticles and their activity against pathogenic bacteria’, Spectrochim. Acta A, Mol. Biomol. Spectrosc., 2012, 91, pp. 23 –29 (doi: 10.1016/j.saa.2012.01.011) [DOI] [PubMed] [Google Scholar]
- 10. Prabhu S. Poulose E.K.: ‘Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects’, Int. Nano Lett., 2012, 2, pp. 1 –10 (doi: 10.1186/2228-5326-2-32) [DOI] [Google Scholar]
- 11. Golinska P. Wypij M. Ingle A.P. et al.: ‘Biogenic synthesis of metal nanoparticles from actinomycetes: biomedical applications and cytotoxicity’, Appl. Microbiol. Biotechnol., 2014, 98, pp. 8083 –8097 (doi: 10.1007/s00253-014-5953-7) [DOI] [PubMed] [Google Scholar]
- 12. Duhan J.S. Gahlawat S.K.: ‘Biogenesis of nanoparticles: a review’, Afr. J. Biotechnol., 2015, 13, pp. 2778 –2785 [Google Scholar]
- 13. Durán N. Marcato P.D. Duran M. et al.: ‘Mechanistic aspects in the biogenic synthesis of extracellular metal nanoparticles by peptides, bacteria, fungi, and plants’, Appl. Microbiol. Biotechnol., 2011, 90, pp. 1609 –1624 (doi: 10.1007/s00253-011-3249-8) [DOI] [PubMed] [Google Scholar]
- 14. El‐Shanshoury A.E.R.R. ElSilk S.E. Ebeid M.E.: ‘Extracellular biosynthesis of silver nanoparticles using Escherichia coli ATCC 8739, Bacillus subtilis ATCC 6633, and Streptococcus thermophilus ESh1 and their antimicrobial activities’, ISRN Nanotechnol., 2011, 2011, pp. 1 –7 (doi: 10.5402/2011/385480) [DOI] [Google Scholar]
- 15. Juibari M.M. Abbasalizadeh S. Jouzani G.S. et al.: ‘Intensified biosynthesis of silver nanoparticles using a native extremophilic Ureibacillus thermosphaericus strain’, Mater. Lett., 2011, 65, pp. 1014 –1017 (doi: 10.1016/j.matlet.2010.12.056) [DOI] [Google Scholar]
- 16. Nanda A. Saravanan M.: ‘Biosynthesis of silver nanoparticles from Staphylococcus aureus and its antimicrobial activity against MRSA and MRSE’, Nanomed. Nanotechnol. Biol. Med., 2009, 5, pp. 452 –456 (doi: 10.1016/j.nano.2009.01.012) [DOI] [PubMed] [Google Scholar]
- 17. Jain D. Kachhwaha S. Jain R. et al.: ‘Novel microbial route to synthesize silver nanoparticles using spore crystal mixture of Bacillus thuringiensis ‘, Ind. J. Exp. Biol., 2010, 48, p. 1152 [PubMed] [Google Scholar]
- 18. Busi S. Rajkumari J. Ranjan B. et al.: ‘Green rapid biogenic synthesis of bioactive silver nanoparticles (AgNPs) using Pseudomonas aeruginosa ’, IET Nanobiotechnol., 2014, 8, pp. 267 –274 (doi: 10.1049/iet-nbt.2013.0059) [DOI] [PubMed] [Google Scholar]
- 19. Sunkar S. Nachiyar C.V.: ‘Biogenesis of antibacterial silver nanoparticles using the endophytic bacterium Bacillus cereus isolated from Garcinia xanthochymus’, Asian Pac. J. Trop. Biomed., 2012, 2, pp. 953 –959 (doi: 10.1016/S2221-1691(13)60006-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhanga Y. Chenga X. Zhanga Y. et al.: ‘Biosynthesis of silver nanoparticles at room temperature using aqueous aloe leaf extract and antibacterial properties’, Colloid Surf. Physicochem. Eng. Aspects, 2013, 423, pp. 63 –68 (doi: 10.1016/j.colsurfa.2013.01.059) [DOI] [Google Scholar]
- 21. Rafie M.H.E. Shaheen T.I. Mohamed A.A. et al.: ‘Bio‐synthesis and applications of silver nanoparticles onto cotton fabrics’, Carbohydr. Polym., 2012, 90, pp. 915 –920 (doi: 10.1016/j.carbpol.2012.06.020) [DOI] [PubMed] [Google Scholar]
- 22. Franci G. Falanga A. Galdiero S. et al.: ‘Silver nanoparticles as potential antibacterial agents’, Molecules, 2015, 20, pp. 8856 –8874 (doi: 10.3390/molecules20058856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Azam A. Ahmed A.S. Oves M. et al.: ‘Size‐dependent antimicrobial properties of CuO nanoparticles against Gram‐positive and negative bacterial strains’, Int. J. Nanomed., 2012, 7, p. 3527 (doi: 10.2147/IJN.S29020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Azam A. Ahmed A.S. Oves M. et al.: ‘Antimicrobial activity of metal oxide nanoparticles against Gram‐positive and Gram‐negative bacteria: a comparative study’, Int. J. Nanomed., 2012, 7, p. 6003 (doi: 10.2147/IJN.S35347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Manke A. Wang L. Rojanasakul Y.: ‘Mechanisms of nanoparticle‐induced oxidative stress and toxicity’, BioMed Res. Int., 2013, 2013, pp. 1 –15 (doi: 10.1155/2013/942916) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Syed S. Zubair A. Frieri M.: ‘Immune response to nanomaterials: implications for medicine and literature review’, Curr. Allergy Asthma Rep., 2013, 13, pp. 50 –57 (doi: 10.1007/s11882-012-0302-3) [DOI] [PubMed] [Google Scholar]
- 27. Martínez‐Gutierrez F. Thi E.P. Silverman J.M. et al.: ‘Antibacterial activity, inflammatory response, coagulation and cytotoxicity effects of silver nanoparticles’, Nanomed. Nanotechnol. Biol. Med., 2012, 8, pp. 328 –336 (doi: 10.1016/j.nano.2011.06.014) [DOI] [PubMed] [Google Scholar]
- 28. Bergey D.H.: ‘Bergey's manual of determinative bacteriology’ (Lippincott Williams & Wilkins, 1994, 9th edn.) Baltimore: Williams & Wilkins [Google Scholar]
- 29. Singh D. Rathod V. Ninganagouda S. et al.: ‘Optimization and characterization of silver nanoparticle by endophytic fungi Penicillium sp. isolated from Curcuma longa (turmeric) and application studies against MDR E. coli and S. aureus ‘, Bioinorg. Chem. Appl., 2014, 2014, pp. 1 –8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Singh R. Wagh P. Wadhwani S. et al.: ‘Synthesis, optimization, and characterization of silver nanoparticles from Acinetobacter calcoaceticus and their enhanced antibacterial activity when combined with antibiotics’, Int. J. Nanomed., 2013, 8, p. 4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qi X. Gunawan P. Xu R. et al.: ‘Cefalexin‐immobilized multi‐walled carbon nanotubes show strong antimicrobial and anti‐adhesion properties’, Chem. Eng. Sci., 2012, 84, pp. 552 –556 (doi: 10.1016/j.ces.2012.08.054) [DOI] [Google Scholar]
- 32. Liao K.H. Lin Y.S. Macosko C.W. et al.: ‘Cytotoxicity of graphene oxide and graphene in human erythrocytes and skin fibroblasts’, ACS Appl. Mater. Interfaces, 2011, 3, pp. 2607 –2615 (doi: 10.1021/am200428v) [DOI] [PubMed] [Google Scholar]
- 33. Link S. El‐Sayed M.A.: ‘Optical properties and ultrafast dynamics of metallic nanocrystals’, Annu. Rev. Phys. Chem., 2003, 54, pp. 331 –366 (doi: 10.1146/annurev.physchem.54.011002.103759) [DOI] [PubMed] [Google Scholar]
- 34. Das V.L. Thomas R. Varghese R.T. et al.: ‘Extracellular synthesis of silver nanoparticles by the Bacillus strain CS 11 isolated from industrialized area’, 3 Biotech, 2014, 4, pp. 121 –126 (doi: 10.1007/s13205-013-0130-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalimuthu K. Babu R.S. Venkataraman D. et al.: ‘Biosynthesis of silver nanocrystals by Bacillus licheniformis ‘, Colloids Surf. B, Biointerfaces, 2008, 65, pp. 150 –153 (doi: 10.1016/j.colsurfb.2008.02.018) [DOI] [PubMed] [Google Scholar]
- 36. Sanghi R. Verma P.: ‘A facile green extracellular biosynthesis of CdS nanoparticles by immobilized fungus’, Chem. Eng. J., 2009, 155, pp. 886 –891 (doi: 10.1016/j.cej.2009.08.006) [DOI] [Google Scholar]
- 37. Sintubin L. De Windt W. Dick J. et al.: ‘Lactic acid bacteria as reducing and capping agent for the fast and efficient production of silver nanoparticles’, Appl. Microbiol. Biotechnol., 2009, 84, pp. 741 –749 (doi: 10.1007/s00253-009-2032-6) [DOI] [PubMed] [Google Scholar]
- 38. Nayak R.R. Pradhan N. Behera D. et al.: ‘Green synthesis of silver nanoparticle by Penicillium purpurogenum NPMF: the process and optimization’, J. Nanoparticle Res., 2011, 13, pp. 3129 –3137 (doi: 10.1007/s11051-010-0208-8) [DOI] [Google Scholar]
- 39. Sarkar S. Jana A.D. Samanta S.K. et al.: ‘Facile synthesis of silver nano particles with highly efficient anti‐microbial property’, Polyhedron, 2007, 26, pp. 4419 –4426 (doi: 10.1016/j.poly.2007.05.056) [DOI] [Google Scholar]
- 40. Gurunathan S. Kalishwaralal K. Vaidyanathan R. et al.: ‘Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli ‘, Colloids Surf. B, Biointerfaces, 2009, 74, pp. 328 –335 (doi: 10.1016/j.colsurfb.2009.07.048) [DOI] [PubMed] [Google Scholar]
- 41. Panyala N.R. Pena‐Mendez E.M. Havel J.: ‘Silver or silver nanoparticles: a hazardous threat to the environment and human health’, J. Appl. Biomed., 2008, 6, pp. 117 –129 [Google Scholar]
- 42. Taheri S.M.: ‘Biosynthesis of quasi‐spherical Ag nanoparticle by P. aeruginosa as a bioreducing agent’, Eur. Phys. J. Appl. Phys., 2011, 56, pp. 1 –4 [Google Scholar]
- 43. Hu C.M.J. Zhang L. Aryal S. et al.: ‘Erythrocyte membrane‐camouflaged polymeric nanoparticles as a biomimetic delivery platform’, Proc. Natl. Acad. Sci., 2011, 108, pp. 10980 –10985 (doi: 10.1073/pnas.1106634108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rodriguez P.L. Harada T. Christian D.A. et al.: ‘Minimal ‘self’ peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles’, Science, 2013, 339, pp. 971 –975 (doi: 10.1126/science.1229568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morones J.R. Elechiguerra J.L. Camacho A. et al.: ‘The bactericidal effect of silver nanoparticles’, Nanotechnology, 2005, 16, p. 2346 (doi: 10.1088/0957-4484/16/10/059) [DOI] [PubMed] [Google Scholar]
- 46. Li W.R. Xie X.B. Shi Q.S. et al.: ‘Antibacterial effect of silver nanoparticles on Staphylococcus aureus ‘, Biometals, 2011, 24, pp. 135 –141 (doi: 10.1007/s10534-010-9381-6) [DOI] [PubMed] [Google Scholar]
- 47. Daima H.K. Selvakannan P.R. Shukla R. et al.: ‘Fine‐tuning the antimicrobial profile of biocompatible gold nanoparticles by sequential surface functionalization using polyoxometalates and lysine’, PLoS ONE, 2013, 8, p. e79676 (doi: 10.1371/journal.pone.0079676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Venkatesan B. Subramanian V. Tumala A. et al.: ‘Rapid synthesis of biocompatible silver nanoparticles using aqueous extract of Rosa damascena petals and evaluation of their anticancer activity’, Asian Pac. J. Trop. Med., 2014, 7, pp. S294 –S300 (doi: 10.1016/S1995-7645(14)60249-2) [DOI] [PubMed] [Google Scholar]


