Figure 3.
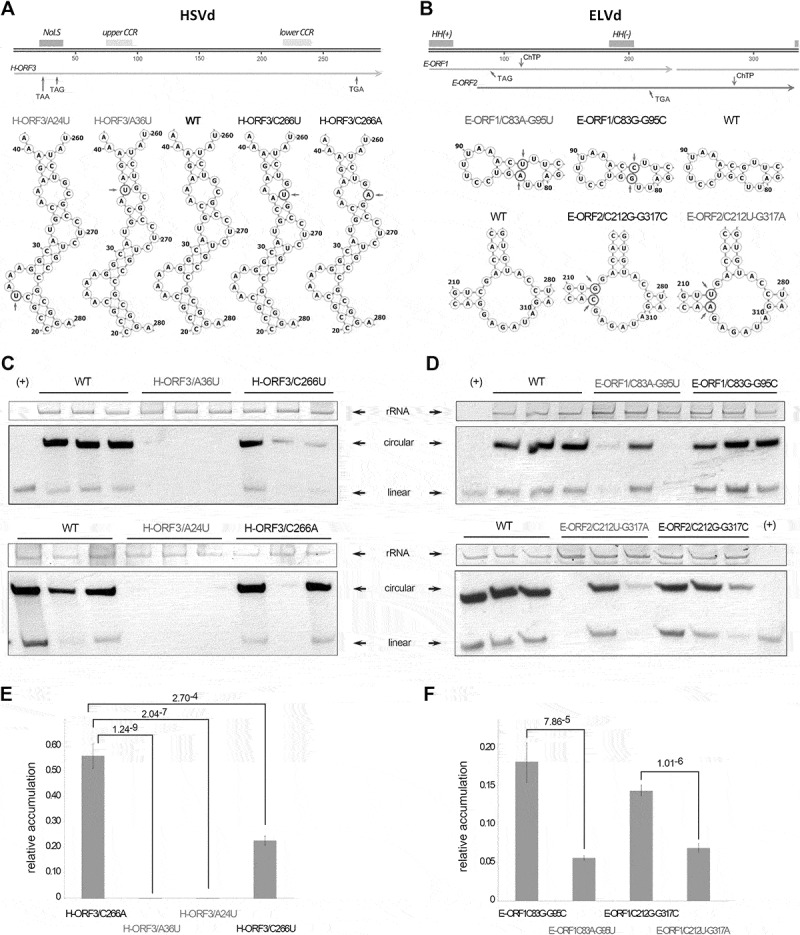
Truncated ORFs affect viroid biological efficiency. Representation of the HSVd (A – upper panel) and ELVd (B – upper panel) linear monomers (plus polarity). The Central Conserved Region (CCR) and the potential Nucleolar Localization Signal (NoLS) (in HSVd) and Hammerhead Ribozyme (HH) (in ELVd) are highlighted (boxes). The position of the nucleotides changed to introduce the stop codons in the conserved ORFs are marked with arrows. Predicted secondary structure of the regions of the HSVd (A – lower panel) and ELVd (B – lower panel) in which were performed the nucleotide substitutions (marked with circles and arrows). WT represents the structure of the unmodified HSVd and ELVd sequences. Representative northern blot hybridization of RNA extracted from cucumber an eggplant inoculated with the HSVd (C) and ELVd (D) mutant-variants. Ribosomal RNA (rRNA) was used as load control. HSVd and ELVd linear transcripts (+) were used as hybridization controls. Relative accumulation of HSVd (E) and ELVd (F) mutant-variants in inoculated plants, estimated by qRT-PCR. The statistical significance was estimated by paired T-tests and the obtained p-values are shown. Error bars represent the standard error values
