Figure 1.
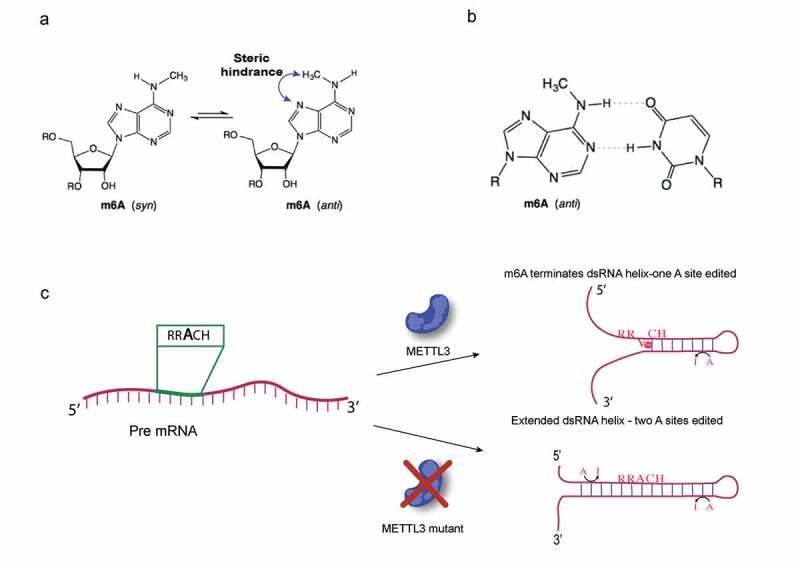
m6A disrupts dsRNA duplex and favours formation of a shorter duplex with m6A stacked on the 5ʹ end of the dsRNA; m6A reduces dsRNA structure and thereby reduces ADAR RNA editing. A. In free nucleotide m6A, the methyl group adopts the relaxed syn conformation to avoid a steric clash with the N7 hydrogen that occurs in the anti conformation of the methyl group. B. In the m6A:U Watson-Crick base pair in dsRNA, the m6A methyl group is forced into the unfavourable, high energy, anti conformation having the steric clash. C. If local dsRNA structure is sufficiently weak, part of the dsRNA melts and m6A, now at the end of a shorter dsRNA duplex, relaxes to the syn conformation. m6A with the methyl in the syn conformation also base stacks on the end of the duplex more favourably than a G would, helping to stabilize the shorter dsRNA. In the presence of the METTL3 writer complex, m6A weakens dsRNA duplexes, either making them shorter or disrupting them entirely if the residual paired stretches are too short. In a Mettl3 mutant or knockdown the unmethylated RRACH sequence can become part of a more stable extended dsRNA helix, increasing the efficiency of ADAR editing at sites already edited in METTL3 wildtype; also, the extended dsRNA may make new sites available for ADAR RNA editing. ADARs require at least 17 base pairs of dsRNA for editing activity
