Abstract
Recently, the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) resulted in Coronavirus Disease 2019 (COVID-19) outbreak. A new SARS-CoV-2 strain is expected to emerge in late 2020, including B.1.1.7. The high transmission rate of SARS-CoV-2 B.1.1.7 has raised public health concerns in several countries. Hence, in this study, we assessed the sequencing of SARS-COV2 to reveals the prevalence of the SARS-CoV-2 Alpha variant (B 1.1.7) in Egypt. We found that the viral transmission of the alpha variant is expanding. Moreover, based on hospitalizations and case fatality rates, there is a potential for increasing severity. There was no effect on susceptibility to Emergency Use Authorization monoclonal antibody treatments. However, there was minimal impact on neutralization by convalescent and post-vaccination sera. Samples have been clustered into the 20D sub clade for the majority of them. The eight samples shown in our study are considered the first recorded samples with the Alpha variant in Egypt. Therefore, The Egyptian government, represented by the Ministry of Health, must take all measures to examine the compatibility of the currently used vaccines with this new strain and the feasibility of the treatment protocol presently used with such strains developed in the Arab Republic of Egypt.
Keywords: SARS-CoV-2, COVID-19, Alpha variant, B 1.1.7, Egypt
Abbreviations: BLAST, Basic Local Alignment Search Tool; BWA, Burrows-Wheeler Aligner software; COVID-19, Coronavirus Disease 2019; Ct, Cycle thresholds; EUS, Emergency Use Authorization; NSP1-16, Non-structural proteins 1 1-6; NTD, N-terminal domain; ORF, Open reading frame; RBM, Receptor binding motif; SARS-CoV2, Severe Acute Respiratory Syndrome Coronavirus 2; VOC, Variant of Concern; VOI, Variant of interest; WHO, World health organization
1. Introduction
Recently, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) 2 resulted in Coronavirus Disease 2019 (COVID-19) outbreak. There have been more than 260 million cases of COVID-19 pandemic since the outbreak, with the mortality impact at roughly 3.9 million (Jamwal et al., 2021). Controlling COVID-19 can be enhanced by vaccination, separating from others socially, general cleanliness, and a large number of diagnostic tests (Voysey et al., 2021; Ferretti et al., 2020).
The genome of SARS-CoV-2 has been estimated to be 29.9 kb (Lu et al., 2020). It is composed of a nucleocapsid protein (N) and an envelope. Envelopes are related with three types of structural proteins: spike protein (S), membrane protein (M), and envelope protein (E). N, S, M, and E are four structural proteins in the SARS-CoV-2 genome, although there are total of 16 non-structural proteins (NSP1–16). Also known as NSP 12, RdRp which plays a key part in the proliferation of (Naqvi et al., 2020).
SARS-CoV-2 was found to be transmissible in roughly 55% of cases, with approximately 30% of people remain asymptomatic (Li et al., 2020). Spike protein receptor binding domain alterations appear to be responsible for the increased transmissibility of novel variations (Cherian et al., 2021; Funk et al., 2021). A significant number of people must be tested in order to detect SARS-CoV-2 infections and, more critically, to identify asymptomatic carriers who may unknowingly infect a huge number of people (Lu et al., 2020). A new SARS-CoV-2 strain is expected to emerge in late 2020, and world health organization (WHO) has classified it as a Variant of Concern or VOI (VOCs). As of now, VOCs include Alpha, Beta, Gamma, Delta, and Epsilon, while VOIs include Epsilon, Zeta, Eta, Theta and Kappa and B.1.427/B.1.429 (Epsilon) (WHO, 2021). There has been viral evolution since the discovery of SARS-CoV-2, however these VOCs have increased transmissibility and severity as well as altered antigenicity, which could have consequences for acquired immunity or efficiency of current vaccinations (Collier et al., 2021; Challen et al., 2021; Madhi et al., 2021; Wang et al., 2021).
The high transmission rate of SARS-CoV-2 B.1.1.7 in the United Kingdom (UK) has raised public health concerns. In addition to the D614G mutation, this variation features eight additional non-synonymous variants in spike: H69-V70, Y144, N501Y, A570D, P681H, T716I, and D1118H. At least three B.1.1.7 spike mutations should be of concern, including the two amino acid deletion in the N-terminal domain (NTD), as well as the N501Y mutation in the receptor binding motif (RBM) and P681H mutation at the Furin cleavage site (P681H) (England, 2020; Rambaut et al., 2020; Funk et al., 2021; Voloch et al., 2021). Interestingly, each of these three mutations can be found in other variants of interest, as well. Mathematical modeling data and epidemiological evidence suggest that this variant is more transmissible than the SARS-CoV-2 variants that were in circulation prior to its introduction; evidence of an increased mortality rate has also been reported, despite initial reports that it was not more pathogenic (England, 2021; Volz et al., 2020).
The first case detected in Egypt with the Alpha variant was recorded in March 2021 (GISAID, 2021). Hence, in this study, we assessed the sequencing of SARS-CoV-2 to reveal the prevalence of SARS-CoV-2 variant B 1.1.7 in Egypt.
2. Material and methods
2.1. Sampling
Oropharyngeal swabs (n = 1235) were taken from patients with suspected COVID-19 in fever hospital of army forces in Egypt. The samples were transported under biosafety requirements (transport bag 95Kpa) in viral transport medium Kit (Cat. no. A48498) at (2–4) C° to the biological prevention department at central chemical laboratories of Almaza in Cairo, Egypt.
2.2. Extraction of RNA and RT-PCR detection
Viral RNA was extracted using QIAamp Viral RNA Mini Kit (Cat. no. 52906). Then, samples were tested for three SARS-CoV-2 genes, i.e., ORF1ab and N protein, using the VIASURE kit (Cat. no.VS-NCO213H) according to the manufacturers' instructions. For each sample, we added 5 μL of extracted RNA to 15 μL of dehydrated master mix. Another tube for positive control was prepared by adding 5 μL of Positive Control to 15 μL of dehydrated master mix. The last tube was prepared for negative control by adding 5 μL of Negative Control to 15 μL of dehydrated master mix. Then, qPCR was performed on an Ariamx thermal cycler (Agilent, Germany), using the following conditions: 45 °C for 15 min, 95 °C for 2 min, 40 cycles of 95 °C for 10 s, and 60 °C for 50 s. Cycle thresholds (Ct) were analyzed using auto-analysis settings, with the threshold lines falling within the exponential phase of the fluorescence curves and above any background signal. We used HEX dye as a fluorescent label for each reaction to determine the Ct value for the internal positive control.
2.3. Library preparation and next-generation sequencing
According to manufacturers' instructions, library preparation was done using the AviSeq COV19 NGS Library Prep kit (Ref. AVG202096). Next-generation sequencing was done by using the Illumina machine (Iseq-100 instrument). RNA was measured on Qubit 2.0 Fluorometer by using Qubit™ RNA HS 100 Assay Kit (Cat.No.Q32852). Then, cDNA was prepared by adding RT Primer Mix DP (BATCH No.020402C). In addition, a multiplex PCR reaction was done to amplify the entire SARS-CoV-2 genome. Digestion was done to remove any nonspecific PCR products. After library preparation, the quality of the library and the amplification of viral RNA were visualized via gel electrophoresis and measured using DNA HS 100 Assay Kit (Cat.No.Q32852).
2.4. Data analysis
FASTA files were generated using Abiomix software. Then Nextclade Tool (version 1.5.2) was used for viral genome alignment, mutation calling, quality checks, phylogenetic placement, and clade assignment. Genome sequencing alignment was done by using Burrows-Wheeler Aligner (BWA) software. Basic Local Alignment Search Tool (BLAST) was used to find similarities between nucleotide and amino acid sequences.
3. Results
3.1. Molecular identification
We used RT-PCR for molecular identification. From April 2020 to July 2021, we collected 1235 COVID-19 positive samples, but only a few hundred models were chosen for sequencing (Cq from 14 to 20).
3.1.1. Sequence data analysis and phylogenetic analysis
Whole Genome sequencing alignment using BWA software resulted in 99 successful high-quality genomes. These sequences were covered the entire genome of reference SARS-CoV-2 (RefSeq: NC_045512.2). Basic Local Alignment Search Tool (BLAST) was used to find similarities between nucleotide and amino acid sequences. Hence, nucleotide identity ranged from 99.8% to 100%, and amino acid identity ranged from 99.7% to 100%.
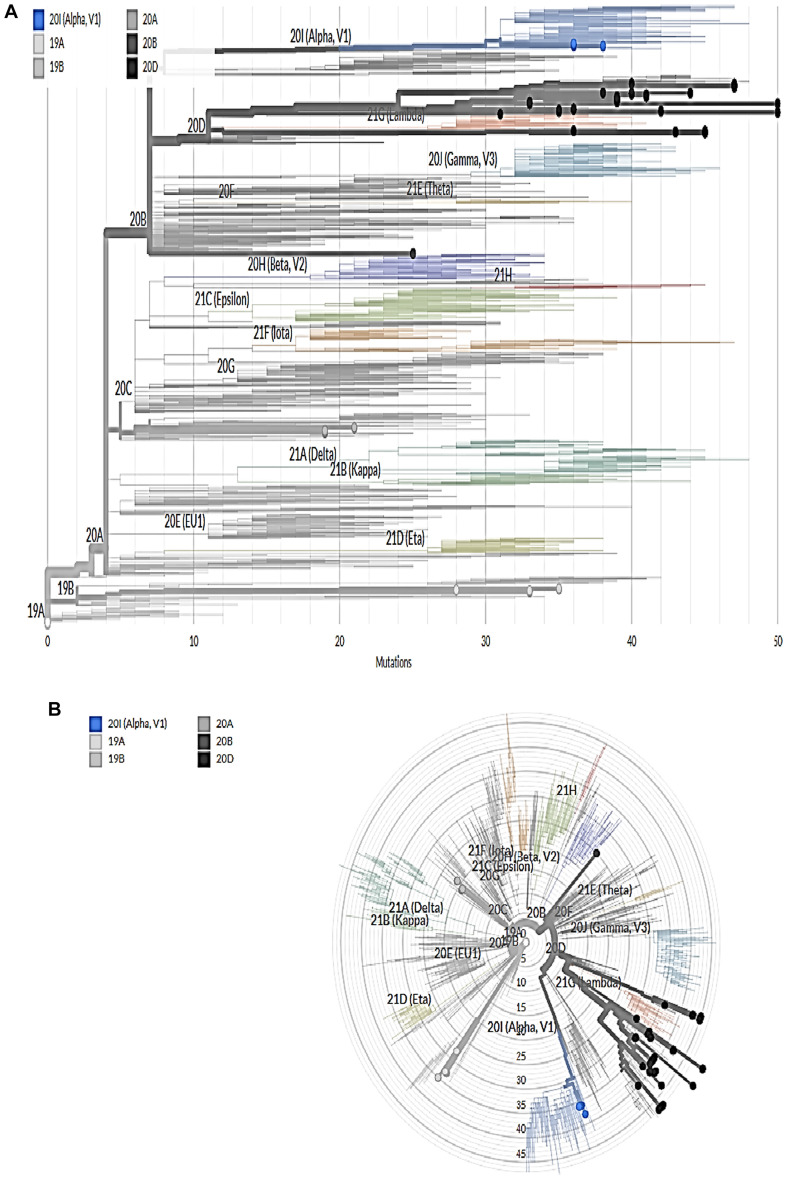
The genetic mutation using Nextclade Tool showed the viral genome, including ORF1a, ORF1b, ORF3a, ORF6, ORF7a, ORF7b, ORF8, S, E, M, and N. [Supplementary file; Figs. A 1–4]. Phylogenetic tree analysis showed six clades in Egypt, as shown in Fig. 1 . These clades were shown in Rectangular and Radial Phylogeny as shown in Fig. 1A and Fig .1B respectively. The most common clade was 20D with 61.6%, and the Alpha variant was 8%.
Fig. 1.
A and B Rectangular and Radial Phylogeny respectively that showing the distribution of our 99 Sequences according to different clades. Using open access server (https://clades.nextstrain.org/tree).
There were about seven sequences clustered as 19A with no mutation in S-gene. However, a single mutation with high quality in ten sequences was assigned as 20A. The clad 20A with D614G amino acids mutations in S-gene was shown in Table 4. Moreover, 11sequences were clustered as 19B with R102S, A292V, N501T, and H655Y amino acids mutations in S-gene. However, two sequences were clustered as 20B with N501T, D614G, and Q677H amino acids mutations in S-gene, as shown in Table 3.
Table 4.
Sequences of 20A and 19A Clades.
| Seq-Name | QC |
Clade | Ns Mut. | S-gene Mut. | non-ACGTN | Ns (missing) | Gaps | Ins. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | M | P | C | F | S | ||||||||
| hCoV-19/Egypt/ARMY-219/2021 EPI_ISL_1936105 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20A | 19 | D614G | 0 | 0 | 0 | 0 |
| hCoV-19/Egypt/ARMY-241/2021 EPI_ISL_1936106 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20A | 19 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/ARMY-228/2021 EPI_ISL_1936107 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20A | 19 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave357/2021 EPI_ISL_1936287 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20A | 19 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave358/2021 EPI_ISL_1936288 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20A | 19 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave359/2021 EPI_ISL_1936289 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20A | 19 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave360/2021 EPI_ISL_1936290 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20A | 19 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/ARMY-212/2021 EPI_ISL_1936141 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20A | 21 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/ARMY-215/2021 EPI_ISL_1936142 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20A | 21 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/ARMY-216/2021 EPI_ISL_1936143 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20A | 21 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-280/2021 EPI_ISL_1936131 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19A | 0 | _ | 0 | 0 | 0 | 0 |
| HCoV-19/Egypt/ARMY-281/2021 EPI_ISL_1936132 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19A | 0 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-282/2021 EPI_ISL_1936133 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19A | 0 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave350/2021 EPI_ISL_1936280 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19A | 0 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave351/2021 EPI_ISL_1936281 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19A | 0 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave352/2021 EPI_ISL_1936282 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19A | 0 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave353/2021 EPI_ISL_1936283 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19A | 0 | 0 | 0 | 0 | 0 | |
N: Missing Data M: Mixed Sites P: Private Mutations C: Mutation Clusters F: Frame shifts S: Stop codons.
Table 3.
Sequences of 20B and 19B Clades.
| Seq-Name | QC |
Clade | Ns Mut. | S-gene Mut. | non-ACGTN | Ns (missing) | Gaps | Ins. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | M | P | C | F | S | ||||||||
| hCoV-19/Egypt/Egypt-Army-Thirdwave313/2021 EPI_ISL_1936252 |
✓ | ✓ | ✘ | ✓ | ✓ | ✓ | 20B | 23 | N501T-D614G-Q677H | 0 | 0 | 0 | 0 |
| hCoV-19/Egypt/Egypt-Army-Thirdwave314/2021 EPI_ISL_1936253 |
✓ | ✓ | ✘ | ✓ | ✓ | ✓ | 20B | 23 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave318/2021 EPI_ISL_1936257 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19B | 25 | R102S-A292V-N501T-H655Y | 0 | 0 | 0 | 0 |
| hCoV-19/Egypt/Egypt-Army-Thirdwave319/2021 EPI_ISL_1936258 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19B | 25 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-273/2021 EPI_ISL_1936128 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19B | 26 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-274/2021 EPI_ISL_1936129 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19B | 26 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-275/2021 EPI_ISL_1936130 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19B | 26 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave347/2021 EPI_ISL_1936277 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19B | 26 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave348/2021 EPI_ISL_19362778 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19B | 26 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave349/2021 EPI_ISL_1936279 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19B | 26 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave310/2021 EPI_ISL_1936249 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19B | 26 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave311/2021 EPI_ISL_1936250 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19B | 26 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave312/2021 EPI_ISL_1936251 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 19B | 26 | 0 | 0 | 0 | 0 | |
N: Missing Data M: Mixed Sites P: Private Mutations C: Mutation Clusters F: Frame shifts S: Stop codons.
About 61 sequences were clustered as 20D with S12F, F59I, W64R, D138Y, W152R, A222V, R346S, L452R, E484K, N501T, T572I, Q613H, D614G, Q677H, D796Y, A871S, A899S, D1163Y, M1237I, 1243S, P1263Q, V1264M, and V1268D amino acids mutations in S-gene as shown in Table 2.
Table 2.
Sequences of 20D Clade.
| Seq-Name | QC |
Clade | Ns Mut. | S-gene Mut. | non-ACGTN | Ns (missing) | Gaps | Ins. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | M | P | C | F | S | ||||||||
| HCoV-19/Egypt/ARMY-231/2021 EPI_ISL_1936134 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 27 | S12F-F59I-W64R-D138Y-W152R-A222V-R346S-L452R-E484K-N501T-T572I-Q613H-D614G-Q677H-D796Y-A871S-A899S-D1163Y-M1237I-C1243S-P1263Q-V1264M-V1268D | 0 | 0 | 0 | 0 |
| HCoV-19/Egypt/ARMY-203/2021 EPI_ISL_1936136 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 27 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-204/2021 EPI_ISL_1936137 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 27 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-205/2021 EPI_ISL_1936138 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 27 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-217/2021 EPI_ISL_1936144 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 27 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave361/2021 EPI_ISL_1936291 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 27 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave362/2021 EPI_ISL_1936292 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 27 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave363/2021 EPI_ISL_1936293 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 27 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave364/2021 EPI_ISL_1936294 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 27 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-267/2021 EPI_ISL_1936127 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 28 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-250/2021 EPI_ISL_1936112 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-251/2021 EPI_ISL_1936113 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-290/2021 EPI_ISL_1936114 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-291/2021 EPI_ISL_1936115 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-252/2021 EPI_ISL_1936116 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-255/2021 EPI_ISL_1936117 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-256/2021 EPI_ISL_1936118 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-257/2021 EPI_ISL_1936119 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-258/2021 EPI_ISL_1936120 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-260/2021 EPI_ISL_1936121 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-261/2021 EPI_ISL_1936122 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-262/2021 EPI_ISL_1936123 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-264/2021 EPI_ISL_1936124 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-265/2021 EPI_ISL_1936125 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-266/2021 EPI_ISL_1936126 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave340/2021 EPI_ISL_1936273 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave342/2021 EPI_ISL_1936274 |
✓ | ✓ | ✘ | ✓ | ✓ | ✓ | 20D | 29 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave320/2021 EPI_ISL_1936259 |
✓ | ✓ | ✘ | ✘ | ✓ | ✓ | 20D | 30 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave321/2021 EPI_ISL_1936260 |
✓ | ✓ | ✘ | ✘ | ✓ | ✓ | 20D | 30 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-208/2021 EPI_ISL_1936102 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 31 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-213/2021 EPI_ISL_1936104 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 32 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-207/2021 EPI_ISL_1936139 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 32 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-211/2021 EPI_ISL_1936140 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 32 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-218/2021 EPI_ISL_1936145 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 32 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave305/2021 EPI_ISL_1936244 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 32 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave306/2021 EPI_ISL_1936245 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 32 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave307/2021 EPI_ISL_1936246 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 32 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-210/2021 EPI_ISL_1936103 |
✓ | ✓ | ✘ | ✓ | ✓ | ✓ | 20D | 34 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave315/2021 EPI_ISL_1936254 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 34 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave316/2021 EPI_ISL_1936255 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 34 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave317/2021 EPI_ISL_1936256 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 34 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave343/2021 EPI_ISL_1936275 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 36 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave300/2021 EPI_ISL_1936240 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave301/2021 EPI_ISL_1936241 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave303/2021 EPI_ISL_1936242 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave304/2021 EPI_ISL_1936243 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave308/2021 EPI_ISL_1936247 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave309/2021 EPI_ISL_1936248 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| HCoV-19/Egypt/Egypt-Army-Thirdwave327/2021 EPI_ISL_1936264 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave328/2021 EPI_ISL_1936265 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave329/2021 EPI_ISL_1936266 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave330/2021 EPI_ISL_1936267 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave331/2021 EPI_ISL_1936268 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave332/2021 EPI_ISL_1936269 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave333/2021 EPI_ISL_1936270 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave334/2021 EPI_ISL_1936271 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave335/2021 EPI_ISL_1936272 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 37 | 0 | 1 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-297/2021 EPI_ISL_1936135 |
✓ | ✓ | ✓ | ✘ | ✓ | ✓ | 20D | 38 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave324/2021 EPI_ISL_1936261 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 39 | 0 | 3 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave325/2021 EPI_ISL_1936262 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 39 | 0 | 3 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave326/2021 EPI_ISL_1936263 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20D | 39 | 0 | 3 | 0 | 0 | |
N: Missing Data M: Mixed Sites P: Private Mutations C: Mutation Clusters F: Frame shifts S: Stop codons.
Finally, eight sequences were clustered as 20I (Alpha, V1) with N501T, D614G, and Q677H amino acid mutations in S-gene, as shown in Table 1 . These eight last sequences were considered the first UK strain detected in Egypt by April 2021 (GISAID, 2021).
Table 1.
Sequences of Alpha variant.
| Seq-Name | QC |
Clade | Ns Mut | S-gene Mut | non-ACGTN | Ns (missing) | Gaps | Ins. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | M | P | C | F | S | ||||||||
| HCoV-19/Egypt/ARMY-239/2021 EPI_ISL_1936108 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20I(Alpha,V1) | 30 | N501Y-A570D-D614G-P681H-T716I-D1118H | 0 | 0 | 0 | 0 |
| HCoV-19/Egypt/ARMY-206/2021 EPI_ISL_1936109 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20I(Alpha,V1) | 30 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-249/2021. EPI_ISL_1936110 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20I(Alpha,V1) | 30 | 0 | 0 | 0 | 0 | |
| HCoV-19/Egypt/ARMY-259/2021 EPI_ISL_1936111 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20I(Alpha,V1) | 30 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave354/2021 EPI_ISL_1936284 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20I(Alpha,V1) | 30 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave355/2021 EPI_ISL_1936285 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20I(Alpha,V1) | 30 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave356/2021 EPI_ISL_1936286 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20I(Alpha,V1) | 30 | 0 | 0 | 0 | 0 | |
| hCoV-19/Egypt/Egypt-Army-Thirdwave344/2021 EPI_ISL_1936276 |
✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 20I(Alpha,V1) | 34 | 0 | 0 | 0 | 0 | |
N: Missing Data M: Mixed Sites P: Private Mutations C: Mutation Clusters F: Frame shifts S: Stop codons.
4. Discussion
According to our study, we found that the viral transmission of the alpha variant is expanding. Moreover, based on hospitalizations and case fatality rates, there is a potential for increasing severity. There was no effect on susceptibility to Emergency Use Authorization (EUA) monoclonal antibody treatments (FDA, 2021). However, there was minimal impact on neutralization by convalescent and post-vaccination sera. Samples have been clustered into the 20D sub clade for the majority of them.
New SARS-CoV-2 variations have emerged in recent months, and their frequency in local populations and even worldwide has shifted fast and repeatedly (Hodcroft et al., 2020; Korber et al., 2020). A number of spike mutations have been found in newer variations, which raises the possibility of immunological escape (Naveca et al., 2021; Tegally et al., 2020).
From the First discovery in England in September 2020, the B.1.1.7 variety has already spread to over 70 countries (Hodcroft et al., 2020). After a mutation was found in the spike protein of a fast-spreading B.1.1.7 strain, it became 56% more transmissible than the earlier SARS-CoV2 strain (Davies et al., 2021a). In South Africa, a new fast-spreading form of SARS-CoV-2 has been identified as B.1.351 virus. SARS-CoV-2 strains of lineage also were recently found in India: B.1.617.1 and B.1.617.2 (Funk et al., 2021). As a result of this new variation, existing control methods are likely to be less successful, and governments may need to take more aggressive efforts to attain the same level of control (Frank, 1996).
Vaccination was affected by the increase in VOC transmission. A rapid and efficient vaccination delivery system is therefore essential for reducing the effects of the pandemic as soon as possible. VOC will also increase herd immunity threshold, which means that future SARS-CoV-2 burden will be greater and more vaccination coverage will be necessary to obtain herd immunity (Davies et al., 2021a).
A previous study reported that there was no significant difference in the neutralization titers between the B.1.1.7 variant and the combination of three Spike mutations. In the B.1.1.7 variant compared to the wild type, the maximum fold change was approximately 6 and the median fold change was approximately 3.85. (IQR 2.68–5.28) (Collier et al., 2021). Another study included 2172 nasal/nasopharyngeal swab samples from 44 counties in California came to this conclusion: B.1.1.7, B.1.351, and P.1 all carry the N501Y mutation. Assays of neutralizing antibody titers from convalescent patients and vaccine recipients showed declines of 4.0 to 6.7-fold and 2.0-fold, respectively (Deng et al., 2021).
According to our study, the genetic mutation showed the viral genome, including ORF1a, ORF1b, ORF3a, ORF6, ORF7a, ORF7b, ORF8, S, E, M, and N. Phylogenetic tree analysis showed six clades in Egypt, as shown in. The most common clade was 20D with 61.6%, and the Alpha variant was 8%. There were about seven sequences clustered as 19A with no mutation in S-gene. However, a single mutation with high quality in ten sequences was assigned as 20A. The clad 20A with D614G amino acids mutations in S-gene was shown in Table 4. Moreover, 11sequences were clustered as 19B with R102S, A292V, N501T, and H655Y amino acids mutations in S-gene. However, two sequences were clustered as 20B with N501T, D614G, and Q677H amino acids mutations in S-gene. About 61 sequences were clustered as 20D with S12F, F59I, W64R, D138Y, W152R, A222V, R346S, L452R, E484K, N501T, T572I, Q613H, D614G, Q677H, D796Y, A871S, A899S, D1163Y, M1237I, 1243S, P1263Q, V1264M, and V1268D amino acids mutations in S-gene.
Finally, eight sequences were clustered as 20I (Alpha, V1) with N501T, D614G, and Q677H amino acid mutations in S-gene. These eight last sequences are considered the first UK strain detected in Egypt by April 2021.
When it comes to B.1.1.7/SGTF, B.1.351 and P.1, there is a higher risk of hospitalization and ICU admission in age groups 60 years in this study. As seen in Germany, B.1.1.7 dominance was associated with an increase in hospitalizations among those under 60 years of age (Funk et al., 2021; Lorbach, 2021). A greater infection rate in younger, school-age age groups was documented in the UK before (Davies et al., 2021b; Gravagnuolo et al., 2021). Denmark has also documented higher hospitalization rates for B.1.1.7 patients (Bager et al., 2021).
When exposed to convalescent sera, variation B.1.1.7 shows only a small reduction in neutralization susceptibility (1.5-fold average) (Muik et al., 2021). When persons receive both doses of both vaccines, neutralization titers have been reported to increase around 10-fold after the second dose, suggesting that a 2-fold decline in neutralization will have no influence on vaccine efficacy (Anderson et al., 2020; Walsh et al., 2020). Recently, it was discovered that the Novavax vaccination was 95,6% effective against the common variant and 85,6% against B.1.1.7 (Callaway and Mallapaty, 2021). In areas where the B.1.1.7 variety is prevalent, receiving the second dose as soon as possible is recommended for optimum efficacy (Shen et al., 2021).
A cautious strategy should be to experiment with variant sequences and conduct post-licensure investigations to detect changes in efficacy between the preexisting and new variants, according to vaccine developers if a vaccine's strain formulation is altered without any other modifications in its content. Licensing authorities may need to clarify the abbreviated marketing paths for the vaccine (Davies et al., 2021a).
Some of our findings have limitations. We can only assess how well the theories suggested are supported by the data. Still, there may be other viable explanations for the recurrence of cases that we did not investigate. Even though more experimental work could shed light on the biological mechanisms behind our observations, given our projections of a rapid future rise in incidence from the Alpha variant without additional control measures, we must consider what new approaches may be required to sufficiently reduce the ongoing transmission of SARS-CoV-2.
5. Conclusion
Viral transmission of the alpha variant is expanding. Moreover, based on hospitalizations and case fatality rates, there is a potential for increasing severity. There was no effect on susceptibility to Emergency Use Authorization monoclonal antibody treatments. However, there was minimal impact on neutralization by convalescent and post-vaccination sera. Samples have been clustered into the 20D sub clade for the majority of them. The eight samples shown in our study are considered the first recorded samples with Alpha variant in Egypt. Therefore, The Egyptian government, represented by the Ministry of Health, must take all measures to examine the compatibility of the currently used vaccines with this new strain and the feasibility of the treatment protocol presently used with such strains developed in the Arab Republic of Egypt.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.105191.
Appendix A. Supplementary data
Supplementary material
References
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383(25):2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bager P., Wohlfahrt J., Fonager J., Albertsen M., Yssing Michaelsen T., Holten Møller C., et al. Increased Risk of Hospitalisation Associated with Infection with SARS-CoV-2 Lineage B.1.1.7 in Denmark. https://ssrn.com/abstract=3792894 Available at SSRN: [DOI] [PMC free article] [PubMed]
- Callaway E., Mallapaty S. Novavax offers first evidence that COVID vaccines protect people against variants. Nature. 2021;590(7844):17. doi: 10.1038/d41586-021-00268-9. [DOI] [PubMed] [Google Scholar]
- Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372 doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian S.P.V., Jadhav S., Yadav P., Gupta N., Das M. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv. 2021 doi: 10.1101/2021.04.22.440932. 04.22.440932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D., Meng B., Ferreira I., Datir R., Temperton N.J., Elmer A., et al. Impact of SARS-CoV-2 B. 1.1. 7 Spike variant on neutralisation potency of sera from individuals vaccinated with Pfizer vaccine BNT162b2. MedRxiv. 2021 doi: 10.1101/2021.01.19.21249840. [DOI] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021 Apr 9;372(6538):eabg3055. doi: 10.1126/science.abg3055. (Epub 2021 Mar 3. PMID: 33658326; PMCID: PMC8128288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Jarvis C.I., Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. MedRxiv. 2021 Mar 9 doi: 10.1101/2021.03.07.21252647. Preprint. 2021.03.07.21252647. (PMID: 33758899; PMCID: PMC7987058) [DOI] [Google Scholar]
- England P.H. Investigation of novel SARS-CoV-2 variant: variant of concern 202012/01. PHE Tech Brief. 2020 [Google Scholar]
- FDA Fact Sheet For Health Care Providers Emergency Use Authorization (Eua) Of Bamlanivimab And Etesevimab 02092021. 2021. https://www.fda.gov/media/151719/download Available from: (Accessed)
- Ferretti L., Wymant C., Kendall M., Zhao L., Nurtay A., Abeler-Dörner L., et al. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368(6491):eabb6936. doi: 10.1126/science.abb6936. (Epub 2020 Mar 31. PMID: 32234805; PMCID: PMC7164555) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.A. Models of parasite virulence. Q. Rev. Biol. 1996;71(1):37–78. doi: 10.1086/419267. (PMID: 8919665) [DOI] [PubMed] [Google Scholar]
- Funk T., Pharris A., Spiteri G., Bundle N., Melidou A., Carr M., et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro Surveill. 2021;26(16) doi: 10.2807/1560-7917.ES.2021.26.16.2100348. pii=2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GISAID Map of tracked variant occurrence. 2021. https://www.gisaid.org/hcov19-variants/ (Accessed)
- Gravagnuolo A.M., Faqih L., Cronshaw C., Wynn J., Klapper P., Wigglesworth M. High throughput diagnostics and dynamic risk assessment of SARS-CoV-2 variants of concern. EBioMedicine. 2021;70 doi: 10.1016/j.ebiom.2021.103540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Zuber M., Nadeau S., Crawford K.H.D., Bloom J.D., Veesler D., et al. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. MedRxiv. 2020 doi: 10.1101/2020.10.25.20219063. [Preprint]. 2021 Mar 24:2020.10.25.20219063. (Update in: Nature. 2021 Jul;595(7869):707–712. PMID: 33269368; PMCID: PMC770918) [DOI] [PubMed] [Google Scholar]
- Jamwal V.L., Kumar N., Bhat R., Jamwal P.S., Singh K., Dogra S., et al. Optimization and validation of RT-LAMP assay for diagnosis of SARS-CoV2 including the globally dominant Delta variant. Virol. J. 2021;18:178. doi: 10.1186/s12985-021-01642-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020 May 1;368(6490):489–493. doi: 10.1126/science.abb3221. (Epub 2020 Mar 16. PMID: 32179701; PMCID: PMC7164387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorbach A. 2021. Nudging im Unternehmen in Bezug auf das Sicherheits-und Gesundheitsverhalten von Arbeitnehmerinnen und Arbeitnehmern unter besonderer Berücksichtigung der Corona-Pandemie. Soc Policy Demand A Work Pap Ser. No 2021/02.https://EconPapers.repec.org/RePEc:sau:sozspd:2102 available at: [DOI] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N Engl J Med. 2021;384(20):1885–1898. doi: 10.1056/NEJMoa2102214. (Epub 2021 Mar 16. PMID: 33725432; PMCID: PMC7993410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.-K., Sänger B., Swanson K.A., Mühl J., Chen W., et al. Neutralization of SARS-CoV-2 lineage B. 1.1. 7 pseudovirus by BNT162b2 vaccine–elicited human sera. Science (80- ) 2021;371(6534):1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866(10):165878. doi: 10.1016/j.bbadis.2020.165878. (Epub 2020 Jun 13. PMID: 32544429; PMCID: PMC7293463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveca F., Nascimento V., Souza V., Corado A., Nascimento F., Silva G., et al. Phylogenetic relationship of SARS-CoV-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the spike protein. Virol Org. 2021;1 [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29(4) doi: 10.1016/j.chom.2021.03.002. 529-539.e3. Epub 2021 Mar 5. PMID: 33705729; PMCID: PMC7934674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. MedRxiv. 2020 doi: 10.1101/2020.12.21.20248640. 12.21.20248640. [DOI] [Google Scholar]
- Voloch C.M., Silva Francisco R., Jr., LGP Almeida, Cardoso C.C., Brustolini O.J., Gerber A.L., et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol. 2021;95(10):e00119–e00121. doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Geidelberg L., Hinsley W.R., The COVID-19 Genomics UK (COG-UK) consortium, et al. Transmission of SARS-CoV-2 Lineage B. 1.1. 7 in England: Insights from linking epidemiological and genetic data. medRxiv. 2020 doi: 10.1101/2020.12.30.20249034. 12.30.20249034; doi. [DOI] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. (Epub 2020 Dec 8. Erratum in: Lancet. 2021 Jan 9;397(10269):98. PMID: 33306989; PMCID: PMC7723445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383(25):2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., et al. Increased Resistance of SARS-CoV-2 Variants B. 1.351 and B. 1.1. 7 to Antibody Neutralization. bioRxiv. 2021 doi: 10.1101/2021.01.25.428137. 01.25.428137. [DOI] [PubMed] [Google Scholar]
- WHO Tracking SARS-CoV-2 variants. 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (Last accessed)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material