FIGURE 2.
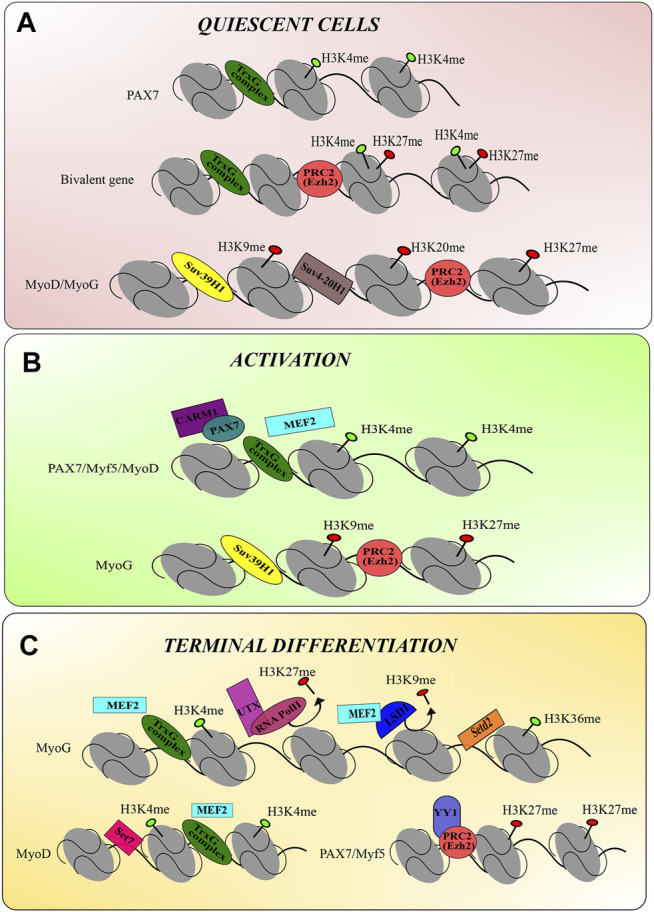
The epigenetic modulation of histone methyltransferases during skeletal muscle regeneration. (A) Epigenetic mechanisms of histone methyltransferases in quiescent stem cells. TrxG-mediated H3K4me catalyses the permissive chromatin of Pax7. MyoD and MyoG gene expression is repressed by H3K27me, H3K9me, and H3K20me via their respective enzymes. Interestingly, the bivalent gene consists of an activating H3K4me mark and the repressive H3K27me, however, H3K27me activity dominates to repress gene transcription during the quiescent state. (B) Epigenetic mechanisms of histone methyltransferases in activating progenitor cells. Upon the activation of stem cells, the permissive chromatin of Pax7, Myf5 and MyoD is catalysed by deposition of H3K4me via TrxG complexes. MEF2 and Carm-Pax7 complex recruits TrxG complexes to establish the permissive chromatin. MyoG remains with the repressive chromatin marks, H3K27me and H3K9me, catalysed by PRC2 and SUV39H1, respectively. (C) Epigenetic mechanisms of histone methyltransferases in during terminal differentiation. The MyoG chromatin permits gene transcription via H3K4me and H3K36me marks by TrxG complexes and Setd2, respectively. UTX and RNA PolII remove the repressive mark H3K27me, and MEF2 recruits LSD1 to remove the H3K9me repressive mark. MyoD gene transcription continues via the activating H3K4me. Pax7 and Myf5 are silenced by recruitment of PRC2 by YY1, which deposits the H3K27me repressive mark.
