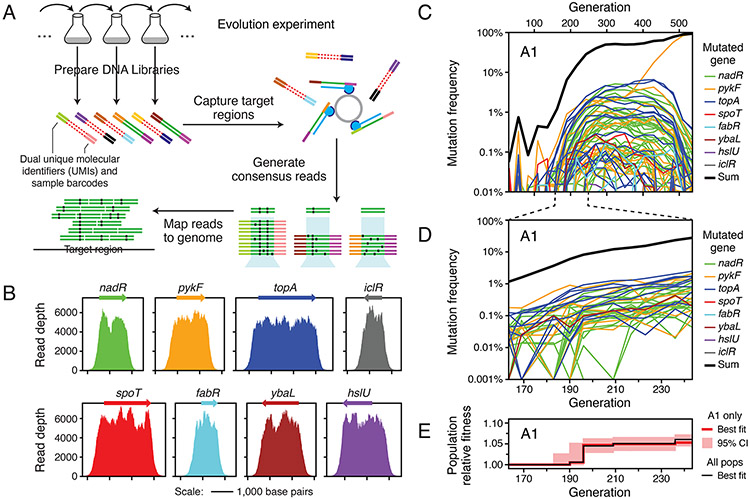
Figure 2. Profiling many beneficial mutations in the first selective sweep by deep sequencing.
(A) Schematic of the deep sequencing approach. Genomic DNA is directly isolated from the E. coli populations and prepared for paired-end Illumina sequencing with sample barcodes and dual UMIs (colored ends attached to red/green double stranded DNA). DNA fragments matching the targeted genome regions (green centers) are captured by probes (blue) bound to magnetic beads and other sequences are washed away (red centers). Reads in pairs that have the same dual unique molecular identifiers, which implies that they were PCR amplified during library preparation from the same original genomic DNA fragment, are used to construct consensus reads to eliminate sequencing errors. Consensus reads are mapped to the reference genome to call sequence variants. (B) Enrichment of reads mapping to eight genes known to be early targets of selection in this environment from the long-term evolution experiment. The final coverage depth of consensus reads in and around these genes is shown for a typical sample (population A7 at 500 generations). (C) Frequency trajectories for mutations in the eight targeted genes as well as the sum total frequency in population A1 over the complete time course of the evolution experiment. When a mutation was not detected at a time point, its trajectory is shown as passing through a frequency of 0.0001% (outside of the plot bounds). (D) Mutation frequency trajectories for population A1 during the selective sweep window from 163 to 243 generations when mutations were first reaching detectable frequencies and outcompeting the ancestral genotype. At time points when a mutation was not detected, its frequency is shown as 0.001% (at the bottom of the plot). (E) Estimated relative fitness of population A1 in each interval between sequenced time points. The frequency trajectories of all beneficial mutations in the initial sweep shown in D were used to jointly estimate the average fitness of the entire population from the deceleration in the rate of increase of the observed mutation trajectories as genotypes with beneficial mutations became common (see Methods). This fitness trajectory fit accounts for all cells in the population, regardless of whether they have a mutation in the targeted genes or elsewhere in the genome. The red line is the maximum likelihood estimate of the population fitness trajectory. The red shading around it shows 95% confidence intervals on this value in each interval. The black line shows the increase in fitness estimated for a consensus model that was jointly fit to all mutations tracked in all six populations. The consensus population fitness trajectory was used when estimating the fitness effects of individual mutations (see Methods).