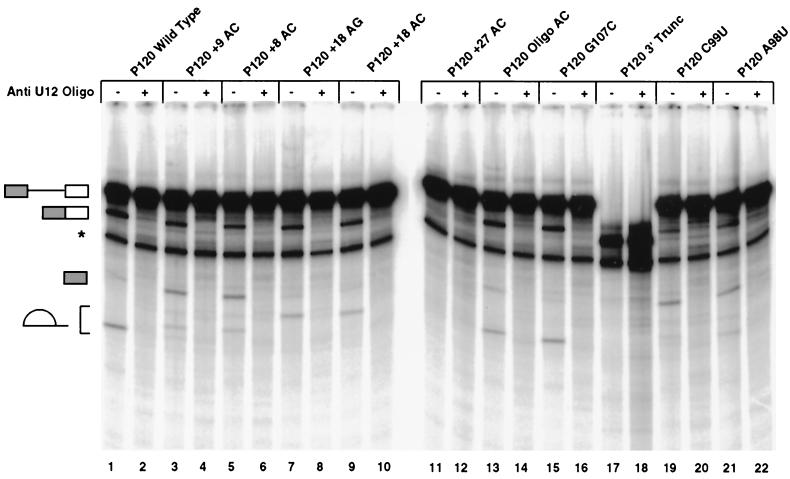
FIG. 6.
In vitro splicing patterns of 3′ splice site constructs. Templates for in vitro transcription of the indicated 3′ splice site constructs were produced by PCR amplification from the minigene constructs tested in vivo. Equal amounts of transcribed precursor RNAs were spliced in vitro. All reactions contained an anti-U2 snRNA 2′-O-methyl oligonucleotide which inhibits U2-dependent splicing. An anti-U12 snRNA 2′-O-methyl oligonucleotide was also added to even-numbered lanes to inhibit U12-dependent splicing. The structures of the various RNA products are shown on the left and correspond, from top to bottom, to the unspliced precursor, spliced exon product, the exon 1 intermediate generated by the first step of splicing, and the excised lariat intron generated by the second step of splicing. The lariat introns from different constructs migrate differently due to the varying length of RNA 3′ of the branch. A degradation product which migrates below the position of spliced exons is labeled with an asterisk. The splicing reactions shown in lanes 17 and 18 used a truncated precursor RNA that terminated 7 nucleotides 3′ of the branch site and thus was missing the 3′ splice site and the downstream exon.