Fig. 1. Schematic overview of single-molecule mechanical fingerprinting with DNA Nanoswitch Calipers (DNCs).
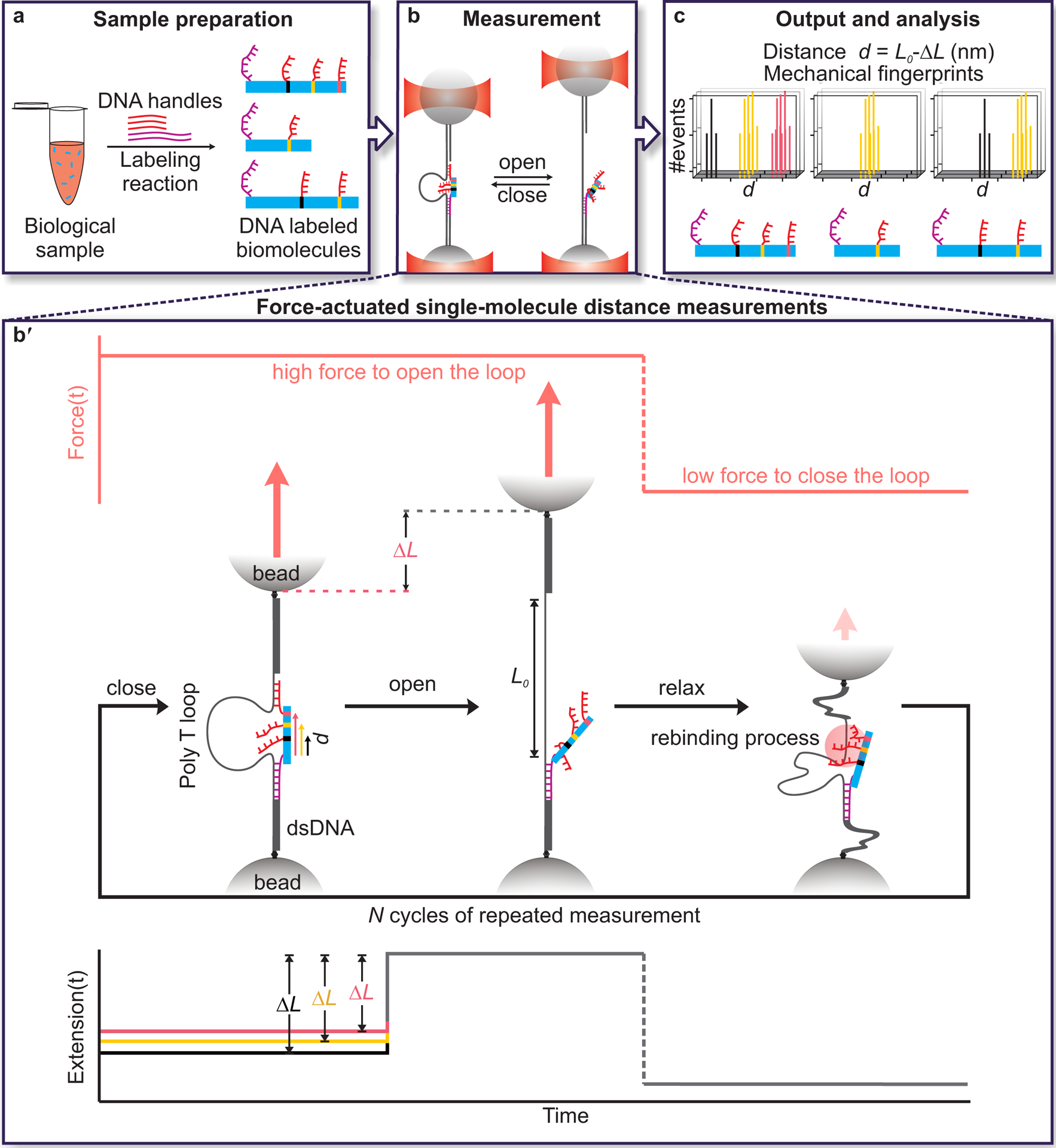
a, Samples are prepared through residue-specific labeling of biomolecules with DNA handles. Here, a long, mechanically strong handle (purple strand) and several shorter, mechanically weak handles (red strands) are attached to each target biomolecule. b, Distances between pairs of handles on target molecules are measured by grabbing and releasing them with the caliper while monitoring the change-in-length (ΔL) between looped and unlooped states. Force-actuation is carried out by tethering the caliper between two optically trapped beads. b′, Detailed view of distance measurements showing the application of force on a DNA Nanoswitch caliper in repeated cycles (top panel), the corresponding changes in conformation of the caliper (middle panel), and the corresponding changes in extension (bottom panel). The increase in length (ΔL) observed when the loop opens during the high-force period depends on the initial distance between handles on the target bridging the loop—the longer the initial distance, the shorter the ΔL. c, Changes in tether length (ΔL) are subtracted from the loop length (L0) to yield absolute distance measurements (d = L0 − ΔL). Multiple measurements are made on each single-molecule target to yield mechanical fingerprints, which can enable the identification of targets in a heterogeneous biological sample. Note that figures are not to scale.
