Figure 1: XBP1 expression signature associates with CRC prognosis.
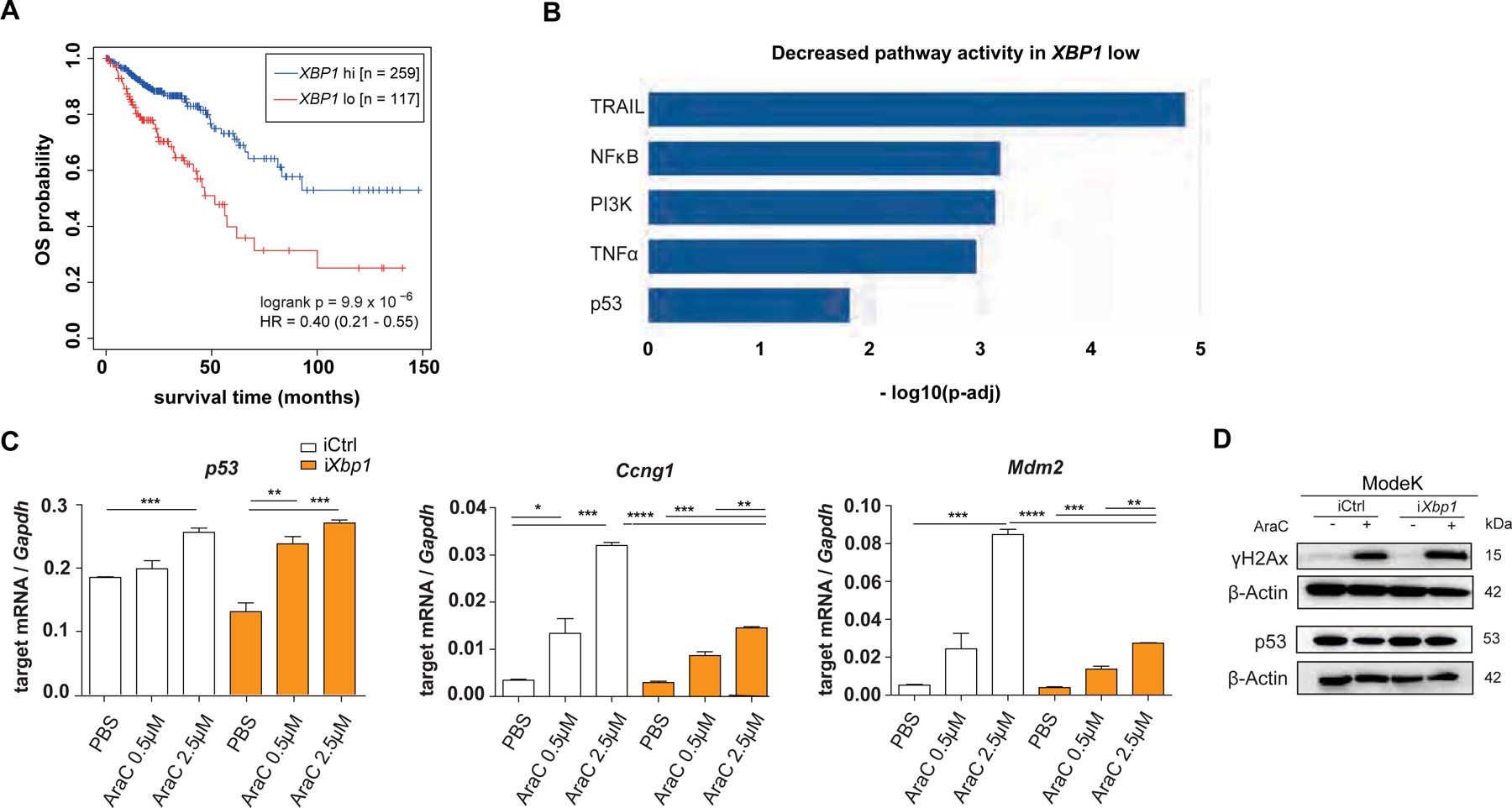
(A) RNA expression data retrieved from the TCGA database (COAD – Colon Adenocarcinoma and READ – Rectum Adenocarcinoma) were used for overall survival (OS) analysis based on high or low XBP1 expression in tumor samples. (B) PROGENy analysis based on pathway enrichment analysis within the same COAD READ dataset and shown as decreased pathway activity in XBP1 low dataset. (C) ModeK cells, treated with PBS, 0.5 μM and 2.5 μM AraC for 24 hours. qRT-PCR analysis of p53-dependent target genes (Sesn2, Ccng1, Mdm2), representative of a minimum of 3 individual experiments with n = 3 technical replicates. Relative mRNA expression is normalized to Gapdh. Data are expressed as mean ± standard error of the mean and significance was determined using an unpaired Student’s t-test. (D) ModeK cells, stimulated with 2.5 μM AraC for 24 hours. Protein lysates were probed against antibodies for p53, γH2Ax and ß-Actin as a loading control on different Western blot membranes, representative of 3 individual experiments. * p < .05, ** p < .01, *** p < .0001, **** p < .00001.
