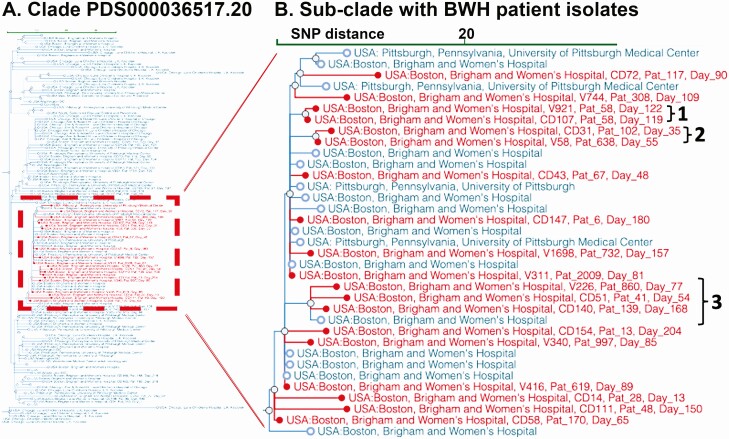
Figure 5.
Subclade selection of Clostridioides difficile genomic clusters by institution and region. The NCBI Pathogen Detection Isolates Browser provides a comparison tool for identifying outbreaks and relating them to other submitted isolate genomes. A, Subclade PDS000036517.20 (135 isolates). B, Subcluster of 35 strains within this clade that includes the largest set of related isolates (n = 19) from the present study (red). Blue entries show 16 prior samples from BWH and 4 from the University of Pittsburgh. The de-identified patient identifier and study day of isolate isolation are overlaid on the NCBI tree. “1” Shows 2 isolates from the same patient; “2” shows 2 closely related isolates that occurred within 20 days but with no identifiable patient spatial overlaps; “3” shows 5 isolates forming a related subclade that includes a sample from BWH submitted 2 years prior to the study (blue). Abbreviations: BWH, Brigham and Women's Hospital; NCBI, National Center for Biotechnology Information; SNP, single nucleotide polymorphism.