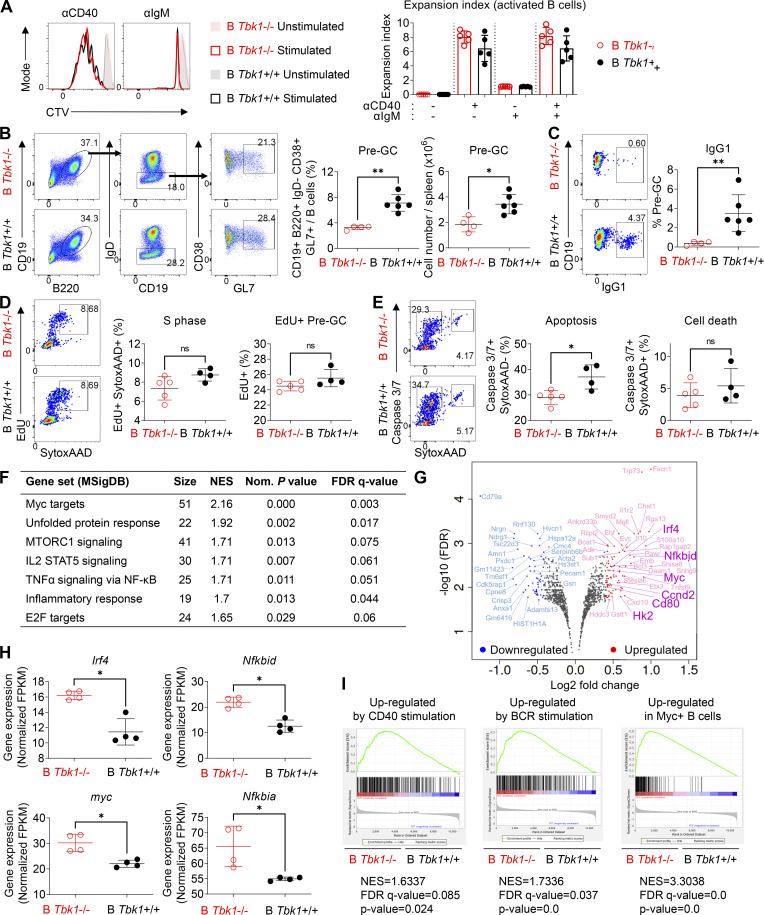
Figure 4.
TBK1 deficiency alters the gene expression in Pre-GC. (A) Representative histograms and expansion index of CTV-stained splenic B cells of day 9 PyNL-infected B Tbk1−/− and B Tbk1+/+ mice stimulated with anti-CD40, anti-IgM, or anti-CD40+ anti-IgM for 4 d analyzed by FACS. n = 4–5 mice/group. (B) Population and cell number of CD38+ GL7+ Pre-GC cells gated from CD19+ B220+ IgD− population in the spleen of B Tbk1−/− and B Tbk1+/+ mice on day 9 after infection with PyNL analyzed by FACS. n = 4–6 mice/group. (C) IgG1 class switching of Pre-GC of B Tbk1−/− and B Tbk1+/+ mice on day 9 after infection with PyNL. n = 4–6 mice/group. (D) Cell cycle analysis of Pre-GC on day 9 after infection with PyNL. n = 4–5 mice/group. (E) Apoptotic cell death analysis of Pre-GC on day 9 after infection with PyNL. n = 4–5 mice/group. (F) Pre-GC isolated from day 9 after infection with PyNL analyzed by RNA-seq. n = 4 mice/group. Gene sets enriched for DEGs between B Tbk1−/− Pre-GC and B Tbk1+/+ Pre-GC analyzed by GSEA using gene signatures from MSigDB Hallmark (false discovery rate [FDR] q value <0.25%; P < 0.05). NES, normalized enrichment score. (G) Volcano plot of DEGs (1.33-fold; P < 0.05) between B Tbk1−/− Pre-GC and B Tbk1+/+ Pre-GC isolated from day 9 after infection with PyNL analyzed by RNA-seq. n = 4 mice/group. (H) Gene expression of Irf4, myc, Nfkbid, and Nfkbia presented as fragments per kilobase of exon per million reads mapped (FPKM) values in RNA-seq of B Tbk1−/− Pre-GC and B Tbk1+/+ Pre-GC. (I) Enrichment of genes up-regulated by CD40 and BCR and expressed in c-Myc+ B cells in B Tbk1−/− Pre-GC compared with B Tbk1+/+ Pre-GC analyzed by GSEA. NES, normalized enrichment score. Each dot represents data from an individual mouse. Data are representative of two (A and C–E) or three independent experiments (B) or from one experiment (F–I). Data are shown as mean ± SD. *, P < 0.05; **, P < 0.01; Mann-Whitney t test (A–E and H). AAD, SYTOX AADvanced.