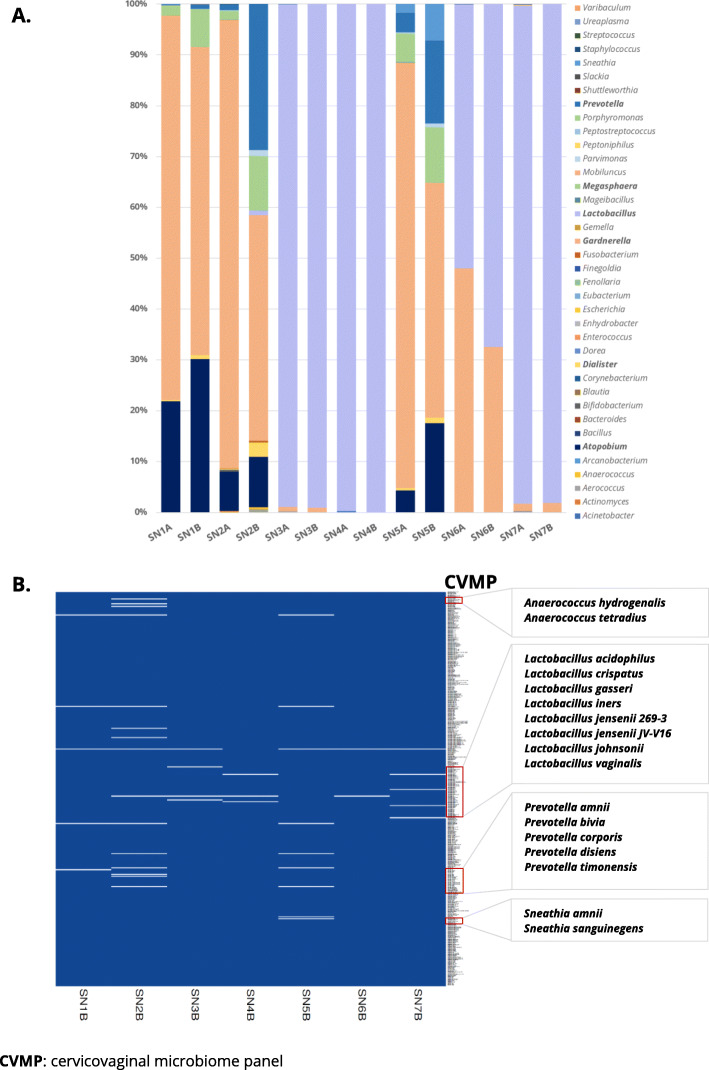
Fig. 4.
CiRNAseq provides genus-level microbiome profiling as 16S rRNA-seq but offers improved taxonomic resolution. A 16S rRNA-seq (SN-A) and CiRNAseq (SN-B) possess similar sequencing capacity when differentiating 31 of the 38 genera analyzed. The methods gave the same results with respect to quantifying the genera Lactobacillus, Gardnerella, Atopobium, and Megasphaera. The microbial composition of samples A and B is similar when analyzed using the two sequencing techniques. Microbial species and isolates URC were summed to show the results at the genus level. B CiRNAseq allows detecting bacteria at high-resolution. The technique suggested 24 different bacterial species, including two species of Anaerococcus, seven Lactobacillus species, five species of Prevotella, and two species of Sneathia. For A, values represent the relative abundances of each microbe in the sample. Bacterial species isolates within our CVMP were considered for display of B