Fig. 6.
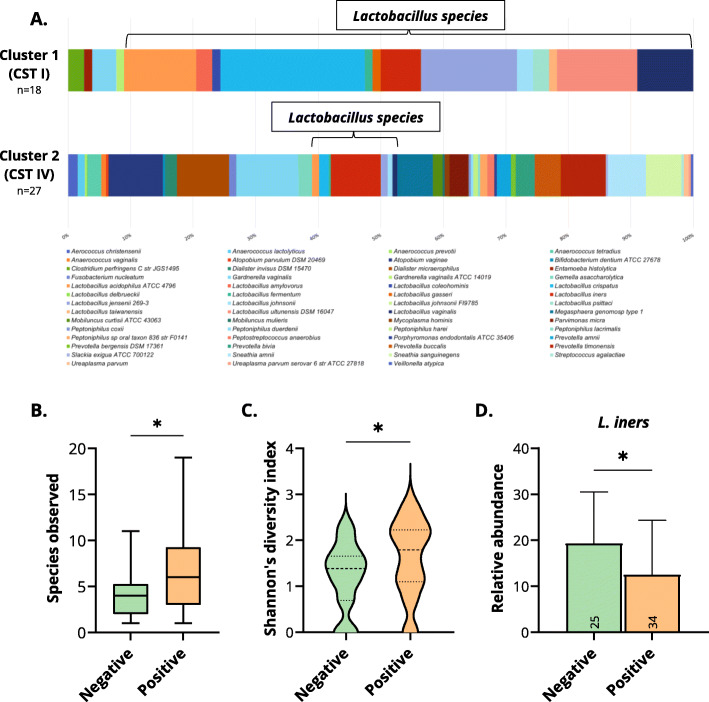
CiRNAseq profiling reveals alterations in the CVM. A The alteration of the microbial diversity at the species level reflects the need for high-resolution sequencing methods. Cluster 1 enriched for CST I and hrHPV-negative women has a less diverse CVM with characteristic Lactobacillus species. In contrast, cluster 2 enriched for CST IV and hrHPV-positive women with CIN2+ contain various microbial species in their microbiome. CST I and IV are derived from the analysis detailed in Fig. 5A. Bacterial isolates from our CVMP were considered for only three species: G. vaginalis, L. johnsonii, and Ureaplasma parvum. Species richness (B) and Shannon’s diversity index (C) further confirm the increase in microbial diversity in hrHPV-positive women. They also demonstrate that hrHPV infections correlate with a rich and diverse CVM. D Using CiRNAseq, we can quantify microbial species within the CVM. L. iners is less abundant in hrHPV-positive women than in hrHPV-negative women, indicating that the progression of hrHPV infections to high-grade cervical lesions is associated with a decreased relative abundance of L. iners. Samples were selected from our cohort of 92 samples. Negative: negative for hrHPV; Positive: positive for hrHPV; *p < 0.05