FIG. 3.
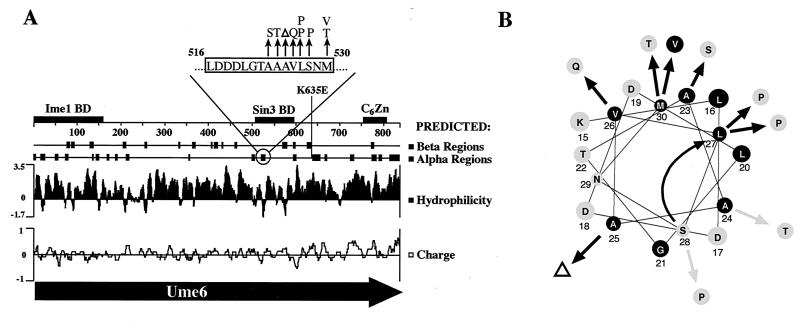
Mutations in UME6 allowing activation by GAD-UME6. (A) Map of UME6 showing locations of mutations and predicted structure. Overall sequence predictions were by the Chou-Fasman method, performed by the Protean sequence analysis program (DNASTAR). The more detailed presentation of the α helix predicted from 516 to 530 (top) is the consensus of four prediction methods (16, 22–24, 52). Mutations are denoted as arrows. Regions known to contain the Ime1 binding domain (BD) (65), Sin3 binding domain (40), and C6Zn DNA binding domain (77) are indicated as boxes. (B) Helical wheel diagram of the predicted α helix (residues 516 to 530 are shown) showing locations of mutations. The curved arrow indicates the shift in position of amino acid 528 resulting from the deletion of amino acid 525.