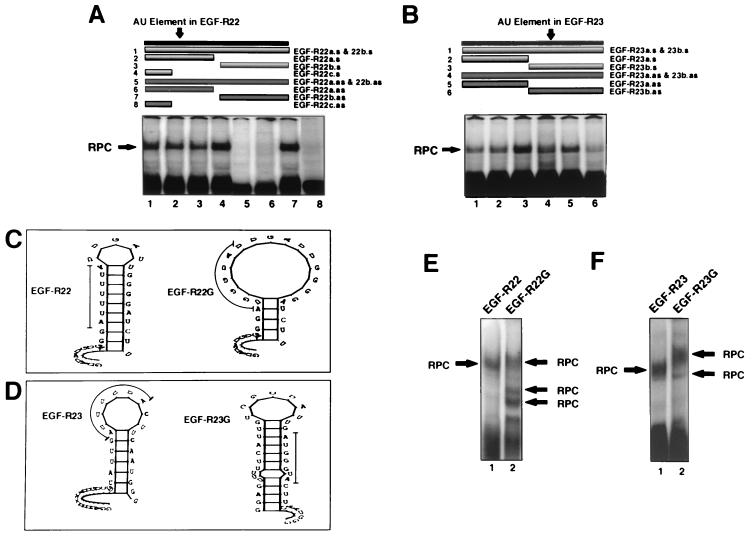
FIG. 9.
Analysis of the EGF-R mRNA binding motif using DNA oligomers and AU-extended pentamer mutants. (A and B) REMSA using MDA-MB-468 extracts and 32P-labeled EGF-R22 (A) or EGF-R23 (B). Before addition of cell extract, a sense or antisense DNA oligomer was incubated with the labeled probe (1:1 molar ratio) for 10 min at 22°C. In panel A, sense oligomers were added to samples in lanes 1 to 4, and antisense oligomers were added to samples in lanes 5 to 8. In panel B, sense oligomers are in lanes 1 to 3, and antisense oligomers are in lanes 4 to 6. The upper portion of each panel illustrates the length and location of each oligomer relative to the labeled probe. The oligomer, probe, and extract mixture was treated as above for REMSA and analyzed by PhosphorImager. The sequence and orientation of each oligonucleotide and probe are shown in Fig. 1D. (C and D) Stem-loop secondary structure plots of EGF-R22 and EGF-R23 and their respective triple-G AU-extended pentamer mutations. The thin line denotes the location of the mutation. The ΔE values for EGF-R22, EGF-R22G, EGF-R23, and EGF-R23G were −3.5, 0.7, −4.0, and 5.7 kJ mol−1, respectively. (E and F) MDA-MB-468 extract was incubated with either 32P-labeled EGF-R22 or EGF-R22G (E) or EGF-R23 or EGF-R23G (F). REMSA was performed as for Fig. 6 and analyzed by PhosphorImager. RPC, RNA-protein complex.