Fig. 2. The terminus-specific recognition of pre-siRNA.
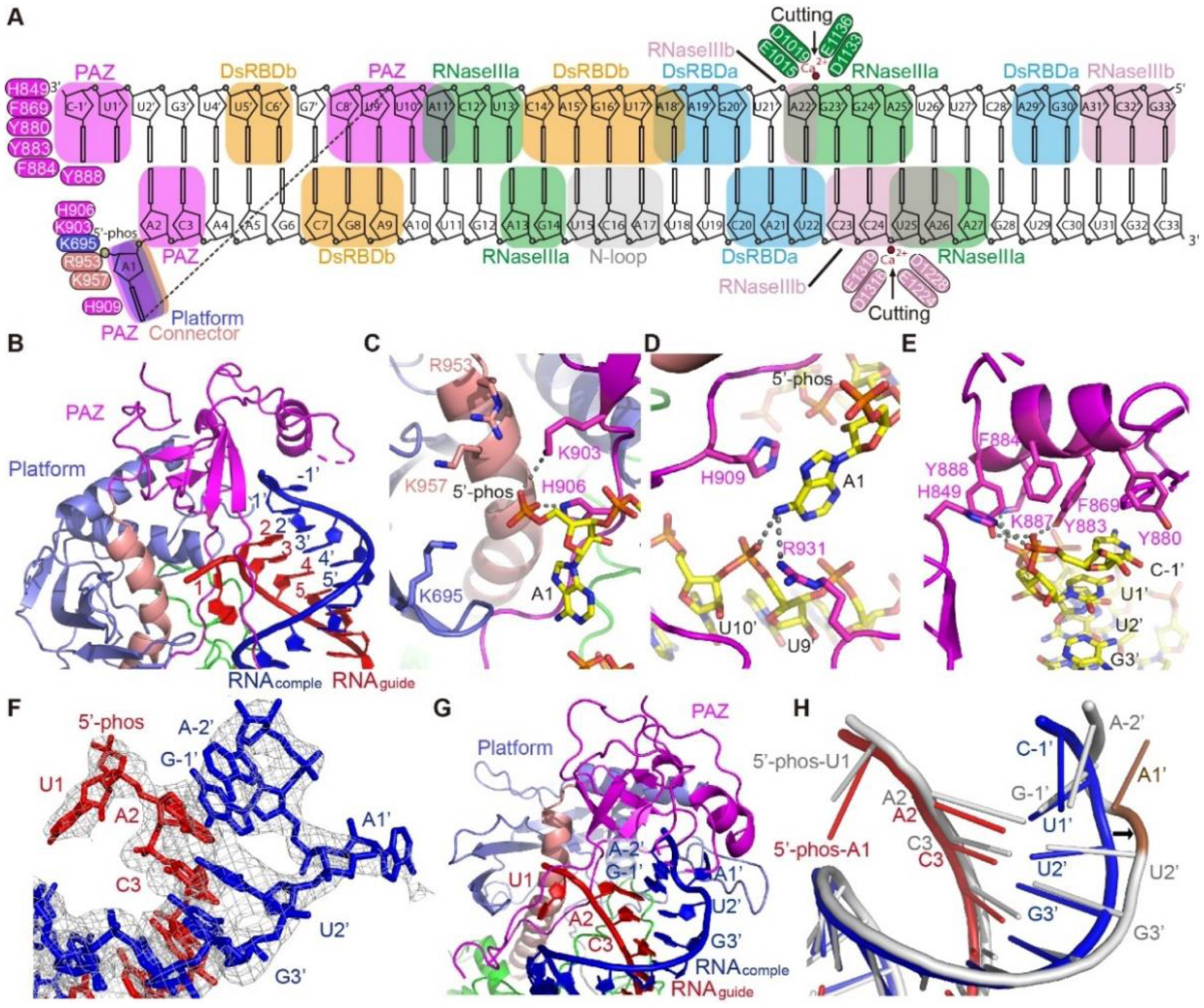
A. Schematic representation of the overall interactions between DCL3 and the pre-siRNA. Key residues are highlighted. The RNA-interacting DCL3 domains are indicated by boxes. B. Overall recognition of the RNA termini by the DCL3 platform-PAZ-connector cassette. The guide strand (RNAguide) and complementary strand (RNAcomple) are highlighted in red and blue, respectively. C. Recognition of the guide strand 5’-phophate group by a positively charged pocket in the platform-PAZ-connector cassette. Interacting residues and hydrogen bonds are highlighted in sticks and dashed-lines, respectively. D. Specific recognition of the A1 base by stacking with DCL3 H909 and hydrogen bonding interactions with DCL3 R931 and the complementary strand. E. An aromatic cap covers and interacts with the last base of the 3’-end of the complementary strand. In addition, the phosphate backbone is recognized by hydrogen bonds. F. Termini of the pre-siRNA with 5’-phosphorylated-U1 on the guide strand and 3’-2-nt overhang on the complementary strand. The cryo-EM map is shown in mesh, showing that both the bases of the U1-A1’ pair were flipped-out. G. Capturing of the RNA termini with 5’-phosphorylated-U1 and 2-nt 3’-overhang by the platform-PAZ-connector cassette. H. Superimposition of the 1-nt 3’-overhang containing RNA duplex (in red and blue) and 2-nt 3’-overhang containing RNA (in silver). The conformational change of RNA is highlighted by the arrow. The flipped out A1’ base allows the G-1’ and A-2’ to fold back and overlap with positions of U1’ and C-1’ of 1-nt-overhang RNA, occupying the same 3’-end position.
