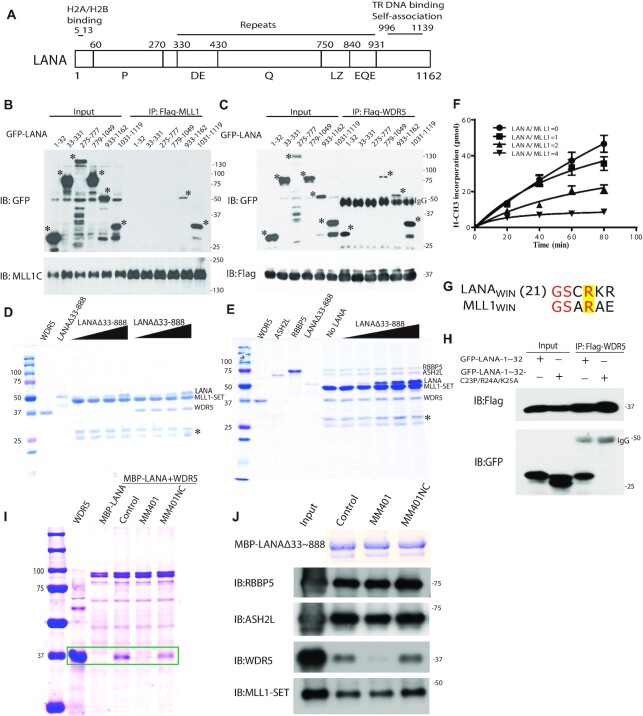
Figure 4.
LANA interacts with, and inhibits, the MLL1 complex. (A) Schematic diagram of LANA. LANA proline rich (P), aspartate and glutamate (DE), glutamine (Q), putative leucine zipper (LZ), glutamate and glutamine (EQE) regions. Residues 5–13 bind histones H2A/H2B (34,77). LANA 996 –1139 self-associates to bind terminal repeat (TR) DNA and independently associates with mitotic chromosomes. (B and C) Co-immunoprecipitation (Co-IP) of GFP-LANA regions with (B) MLL1 or (C) WDR5 after expression in 293T cells. IPs were performed using anti-Flag antibody. Asterisks denote GFP-LANA fusions. (D) LANAΔ33–888, or WDR5 (in lanes at right) co-precipitation with GST-MLL1-SET (E) ASH2L, RbBP5, WDR5 and LANAΔ33–888 co-precipitation with GST-MLL1-SET. (D, E) Triangles at top indicate increasing LANAΔ33–888 and asterisk indicates degradation products. (F) Histone methyltransferase activity of the MLL1 complex after incubation with increasing amounts of LANA. Results are the means of three independent experiments. Standard deviation is shown. (G) Sequence alignment of MLL1WIN with LANAWIN starting at LANA residue 21. Yellow highlight indicates the conserved arginine. (H) Co-IP of GFP-LANA-1–32 or GFP LANA-1–32 containing C23P/R24A/K25A substitutions with Flag-WDR5 after co-expression in 293T cells. (I) Co-precipitation of WDR5 with MBP-LANAΔ33–888 in the presence or absence MM401 or enantiomer control. Two left lanes, WDR5 or MBP-LANAΔ33–888 alone. Box encloses WDR5 bands. (J) ASH2L, RbBP5, WDR5, or MLL1-SET co-precipitation with MBP-LANAΔ33–888 after incubation in the presence or absence MM401 or enantiomer control. Proteins were detected by immunoblot except MBP-LANAΔ33–888, which was detected by Coomassie blue. (D, E, I) Proteins were detected with Coomassie blue.