Table 2.
Top-ranked SNPs for BMI from random effects fine-mapping (AnnoRE) and GWAS analysis in each of the loci with AnnoRE results attaining locus-wide significance
| SNP | Chr: position | Nearest gene | LD 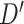 and and 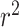
|
MAF | GWAS 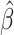
|
GWA.pval | AnnoRE 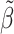
|
Wald scores | AnnoRE pval | GWA rank | AnnoRE rank |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1477196 | 16:53808258 | FTO | 1.0 | 0.28 | −0.058 | 3.22E−72 | −0.147 | 348.049 | 1.13E−77 | 50 | 1 |
| rs1558902 | 16:53803574 | 0.41 | 0.45 | 0.082 | 7.51E−153 | 0.014 | 2.930 | 0.087 | 1 | 31 | |
| rs34358 | 5:74965122 | ANKDD1B | 0.96 | 0.35 | 0.023 | 2.31E−12 | 0.037 | 37.200 | 1.07E−09 | 11 | 1 |
| rs2112347 | 5:75015242 | 0.87 | 0.38 | −0.026 | 6.19E−17 | −0.001 | 0.368 | 0.544 | 1 | 56 | |
| rs988748 | 11:27724745 | BDNF | 0.99 | 0.22 | −0.041 | 5.90E−23 | −0.027 | 33.935 | 5.70E−09 | 16 | 1 |
| rs11030104 | 11:27684517 | 0.89 | 0.20 | 0.041 | 5.56E−28 | 0.003 | 0.729 | 0.393 | 1 | 43 | |
| rs11676272 | 2:25141538 | ADCY3 | 1.0 | 0.48 | 0.032 | 1.12E−21 | 0.031 | 28.952 | 7.42E−08 | 3 | 1 |
| rs10182181 | 2:25150296 | 0.99 | 0.50 | −0.031 | 8.78E−24 | −0.009 | 5.028 | 0.025 | 1 | 4 | |
| rs1048932 | 11:115044850 | CADM1 | 0.90 | 0.50 | −0.019 | 9.43E−10 | −0.019 | 17.367 | 3.08E−05 | 2 | 1 |
| rs12286929 | 11:115022404 | 0.57 | 0.43 | 0.022 | 1.31E−12 | 0.002 | 1.884 | 0.170 | 1 | 7 | |
| rs1800437 | 19:46181392 | QPCTL | 0.99 | 0.18 | 0.035 | 1.73E−17 | 0.022 | 12.912 | 3.27E−04 | 3 | 1 |
| rs2287019 | 19:46202172 | 0.89 | 0.15 | 0.036 | 4.59E−18 | 0.002 | 1.159 | 0.282 | 1 | 10 |
LD measures D′ and 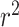 quantify association between the top SNPs identified by the two methods.
quantify association between the top SNPs identified by the two methods.
