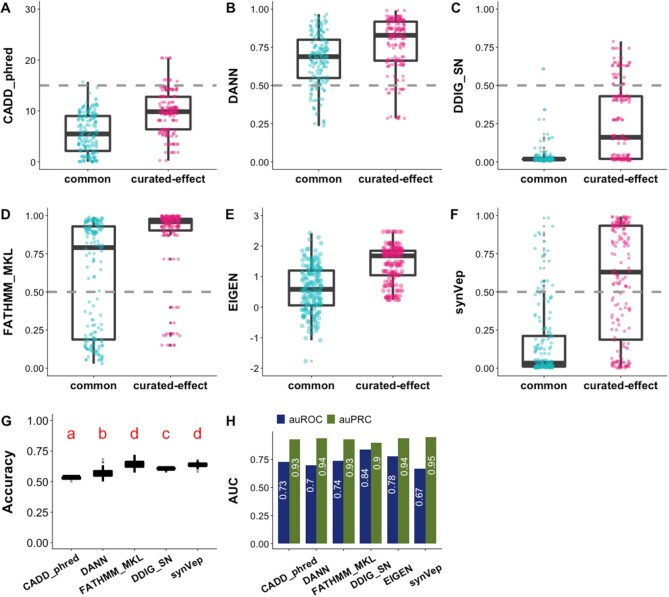
Figure 3.
Predictor performance on common vs. curated-effect sSNVs. Panels A–F show the differential predictions on sets of curated-effect (n = 170) and common sSNVs (randomly selected n = 170) for CADD (phred-like scaled scores), DANN, DDIG-SN, FATHMM-MKL, EIGEN, and synVep, respectively. Gray line indicates scoring cutoff suggested by tool authors. Neither the common set nor the curated-effect set were included in synVep training. Permutation tests show that all predictors give significantly different scores between the effect and common variant sets in every iteration, except for DANN where 11 of 100 comparisons were not significant (P-value > 0.05 after Bonferroni correction). Panel G reports two-class predictor accuracy (Equation 4) on resampled data (100 resampling sets; common set is down-sampled to match the number of curated-effect variants). Predictors with different red letters indicate significant difference by ANOVA test and Tukey's procedure; e.g. CADD’s ‘a’ and DANN’s ‘b’ indicate that CADD’s and DANN’s mean accuracies are significantly different. Panel H reports the performance (auROC and effect auPRC) of each predictor on the left-out common (negative; n = 9274) and curated-effect (positive; n = 170) sSNVs. FATHM, DDIG, and EIGEN auROC and auPRC are significantly different from synVep's (P-value < 0.05; Methods). Note that the performance comparisons here are limited as each predictor targets a different effect (e.g. pathogenicity vs molecular effect) and some methods have used our test set in training.