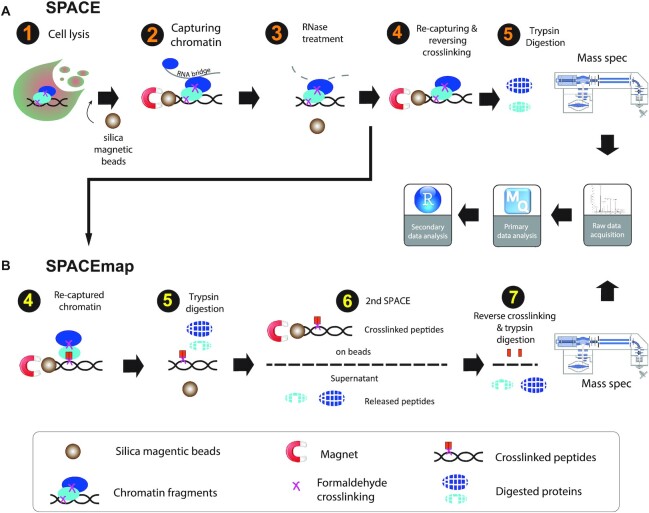
Figure 1.
Overview of SPACE and SPACEmap. (A) In SPACE, 1: Cells are crosslinked by 1% formaldehyde, and resuspended in the lysis buffer containing guanidinium, and iso-propanol. Then silica magnetic beads are added to the lysate. 2: Chromatin binds to the magnetic beads and is separated from the lysate. The beads are washed with lysis buffer and ethanol. 4: Chromatin is eluted by sonication and is treated with RNase A. 4: Chromatin is captured again on the beads to be washed again with ethanol and Acetonitrile. Then the crosslinking is reversed, and trypsin/LysC are added to digest the chromatin-associated proteins on the beads. (B) In SPACEmap, chromatin is recaptured in step 4, however, the crosslinking is not reversed. 5: trypsin is added to digest the chromatin-associated proteins. 6: using another round of SPACE released peptides are separated from crosslinked peptides. Both crosslinked fractions and released fractions are injected to the mass spec to be compared quantitatively. After mass spectrometry and data acquisition, the raw files are analysed by MaxQuant to identify and quantify the proteins. Further statistical, domain and GO analysis are performed using R in RStudio.