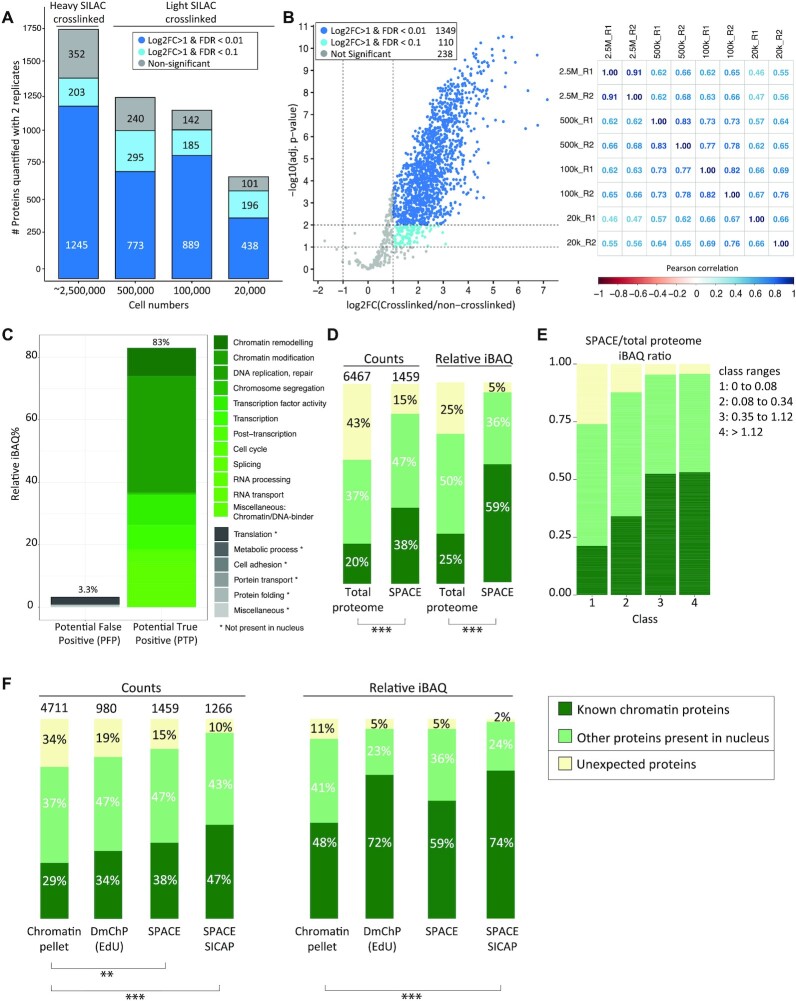
Figure 2.
Chromatin composition in mES cells identified by SPACE. (A) SPACE experiments were carried out by varying number of cells. Each experiment was repeated twice. The bars show the proteins quantified by both replicates using each cell number. The dark blue stacks are highly significantly enriched in comparison to the non-crosslinked control (adj. P-value < 0.01 and log2FC > 1). The blue stacks are significantly enriched in comparison to the non-crosslinked control (adj. P-value < 0.1 and log2FC > 1). The grey stacks are not significantly enriched. (B) All the experiments were integrated, and proteins quantified by at least 1 heavy SILAC crosslinked (forward experiments) and 1 light SILAC crosslinked (reverse experiments) were considered for statistical analysis. The volcano plot shows the proteins that are very significantly enriched in comparison to the non-crosslinked controls with (adj. P-value < 0.01 and log2FC > 1), proteins that are significantly enriched in comparison to the non-crosslinked controls (adj. P-value < 0.1 and log2FC > 1) and proteins that are not significantly enriched with dark blue, blue, and grey, respectively. The matrix shows the Pearson correlations coefficient among the experiments. (C) The enriched proteins by SPACE were categorized based on their biological processes into potential true positive and potential false positive. (D) The enriched proteins were categorized into three groups: 1: ‘known DNA or chromatin binding proteins’ (dark green), 2: ‘Other proteins present in nucleus’ (pale green), 3: Proteins that do not fall into the previous categories are so-called ‘unexpected’ (yellow). The left two bars compare protein counts between the total proteome of mES cells and SPACE. The right two bars compare the relative iBAQ of the proteins. The total proteome data was obtained from published data (20), and re-analysed. Fisher's exact test was used to show the statistical differences. (E) SPACE/total proteome iBAQ ratios for each protein was calculated. The proteins were classified into four equal groups based their ratios. The frequency of 1: ‘known DNA/chromatin-binders’ (dark green), 2: ‘proteins present in Nucleus’ (pale green) and 3: ‘unexpected’ proteins (yellow), was shown in each class. (F) Chromatin pelleting, DmChP, SPACE and SPACE-SICAP results of mES cells were compared based on the protein counts and relative iBAQ of the enriched proteins. Chromatin pelleting (21) and DmChP (17) data were obtained from published data. The enriched proteins were categorized into 3 groups, as mentioned previously in panel B. Fisher's exact test was used to show the statistical differences: *** P-value < 0.001, ** P-value ≤ 0.01 and * P-value ≤ 0.05.