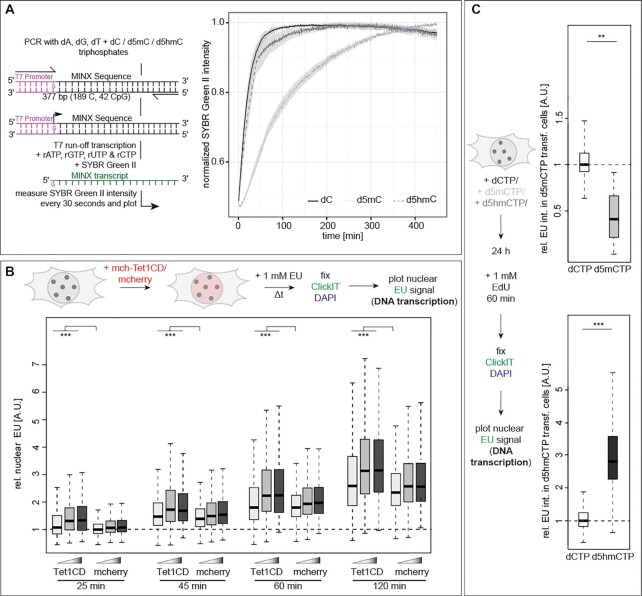
Figure 4.
Effect of cytosine modifications on RNA polymerase activity. (A) Experimental setup for in vitro T7 run-off transcription. Template DNA contained 100% cytosine (dC), methylcytosine (d5mC) or hydroxymethylcytosine (d5hmC) and T7 promoter sequence was unmodified. Normalized mean SYBR green II intensities (± standard deviation), i.e., the amount of RNA transcripts, over time are shown. (B) Schematic representation of the experimental setup for in vivo ribonucleotide incorporation analysis. Cells were incubated with 1 mM EU 24 h after transfection with the constructs as indicated, EU was detected and the normalized nuclear EU sum intensities are plotted. (C) Schematic representation of the experimental setup for in vivo ribonucleotide incorporation analysis in dCTP, d5mCTP or d5hmCTP transfected cells. Cells were incubated with 1 mM EU 24 h after transfection with the nucleotides as indicated, EU was detected and the normalized nuclear EU sum intensities are plotted. Independent experiments were done twice (A and C) or in triplicates (B). All boxes and whiskers are as in Figure 2 and P and n-values are summarized in Supplementary Table S7; ** P < 0.005 and *** P < 0.001.