Figure 3.
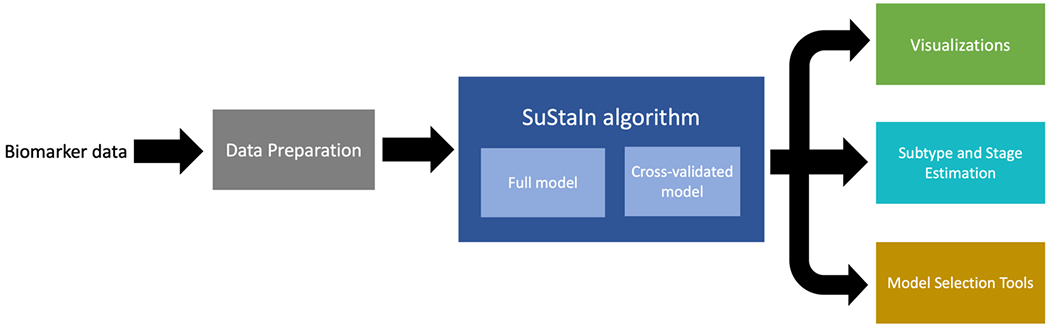
Depiction of the major functionalities of pySuStaIn, as a sequence of operations that begins with an input biomarker data matrix, followed by a data preparation step that depends on the chosen data likelihood, then the SuStaIn algorithm run on both full and cross-validated data and finally a set of outputs consisting of: (i) visualizations of the inferred models; (ii) estimates of the most likely subtype and stage for training and test subjects; and (iii) a set of model selection tools.
