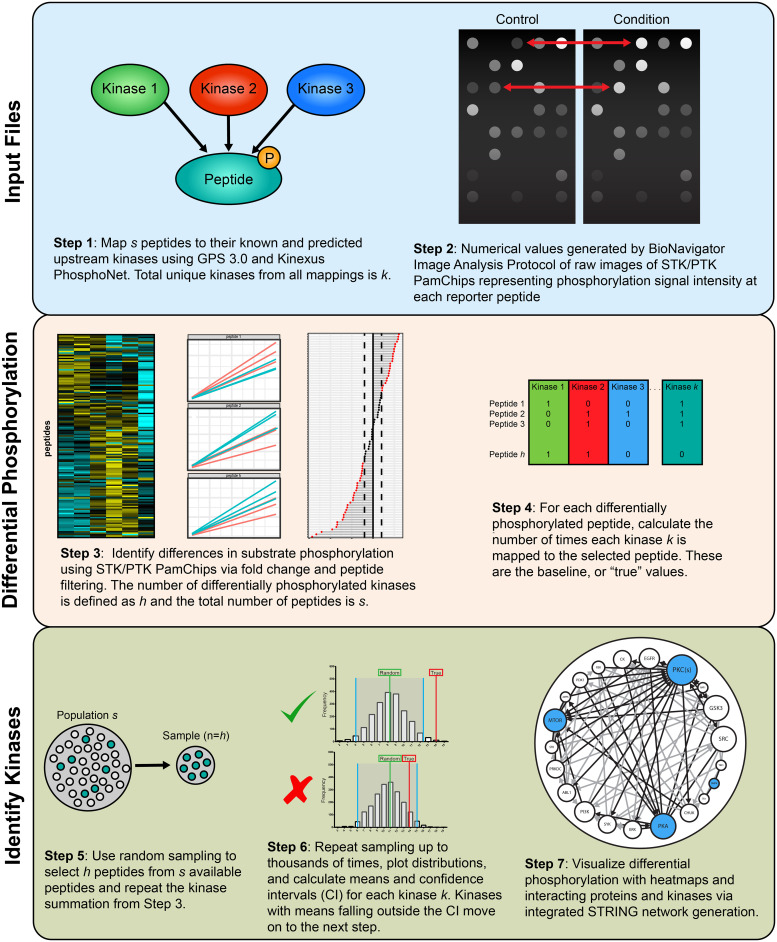
Fig 1. Workflow overview illustrating the primary steps of the KRSA pipeline.
The “Input Files” section outlines the initial input files for KRSA, including the raw kinome array data and the peptide-kinase association file as well as the initial filtering step in KRSA. The “Identify Kinases” section describes the random sampling and distribution evaluation methods used to identify differentially active kinases. Finally, the “Expand and Validate” portion of the figure shows the kinase network generation step of KRSA and confirmation experiments that can be used to validate the predictions from KRSA.