Figure 1.
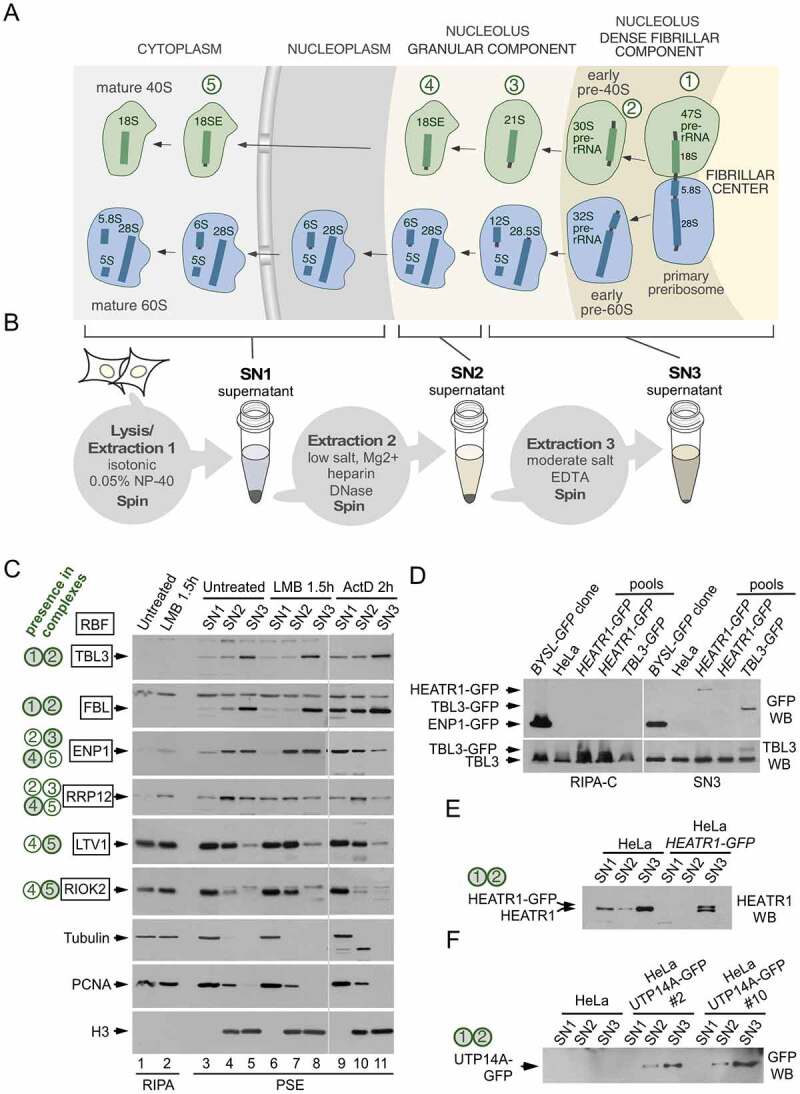
Fractionation of human preribosomes using the PSE extraction method. (a) Simplified scheme of the human ribosome biogenesis pathway showing 40S (green) and 60S (blue) preribosomal particles at different stages of maturation as they travel from the nucleolus to the cytoplasm. The names of the pre-rRNAs species at each stage are indicated. The nucleolus exhibits a tripartite organization that reflects the directionality of preribosome assembly and maturation. The sites of active RNA polymerase I transcription are at the interface between the fibrillar center and the dense fibrillar component, the early processing of the pre-rRNAs occurs in the dense fibrillar component and the late processing in the granular component. The primary transcript (47S pre-rRNA) contains the sequences of the 18S, 5.8S and 28S rRNAs. The earliest pre-40S particles, containing the 47S and 30S pre-rRNAs, are formed in the dense fibrillar component. Five pre-40S maturation stages are indicated with circled numbers. (b) Scheme of the PSE method showing the preribosome pools that are solubilized in each step. There is an additional 18S-E-containing particle (in between stages 3 and 4) that is solubilized in the SN3 fraction (not shown for simplicity). (c) Western blot analyses showing the contents of different RBFs in the SN1, SN2 and SN3 fractions obtained with the PSE method. PSE extraction was performed on HeLa cells normally growing (lanes 3 to 5), and after treatment with LMB (lanes 6 to 8) or ActD (lanes 9 to 11). Equivalent amounts of whole cell lysates prepared with RIPA buffer were analysed in parallel for comparison (lanes 1 and 2). Control proteins, unrelated to the ribosome synthesis route, include tubulin and proliferating cell nuclear antigen (extracted in the SN1 fraction) and histone H3 (extracted in the SN2 and SN3 fractions). The 40S preribosomes that contain each RBF are indicated on the left (green-filled circles are preribosome stages that mostly contain the RBF, open circles are preribosome stages in which only a subpool of particles contain the RBF). A summary of the relevant bibliography and data about the subcellular localization, loss-of-function phenotypes and hierarchy of incorporation into preribosomes of these RBFs can be found in the supplementary information of reference [24]. (d) Western blot analyses of lysates from cell pools that were subject to CRISPR editing to fuse GFP to either HEATR1 or TBL3. Panels on the left correspond to lysate samples prepared with RIPA-C buffer and panels on the right correspond to SN3 fractions obtained with the PSE method of the same pools. The presence of cells expressing the GFP-fusions in some pools is detected in the SN3 but not in the RIPA-C preparations. The parental HeLa cell line and a HeLa-derived cell line that expresses ENP1-GFP (BYSL-GFP clone, described in reference 24) were used as controls. Note that ENP1 is also known as BYSTIN (BYSL is the name of the human gene). (e, f) Western blot analyses showing the PSE fractionation profiles of HEATR1-GFP and UTP14A-GFP proteins endogenously expressed in HeLa-derived cell lines generated by CRISPR editing. Data demonstrating the functionality of the fusion proteins are shown in supplementary figures S1 and S2. HEATR1 and UTP14A are known components of 90S preribosomes in yeast (equivalent to the human particles in stages 1 and 2)
