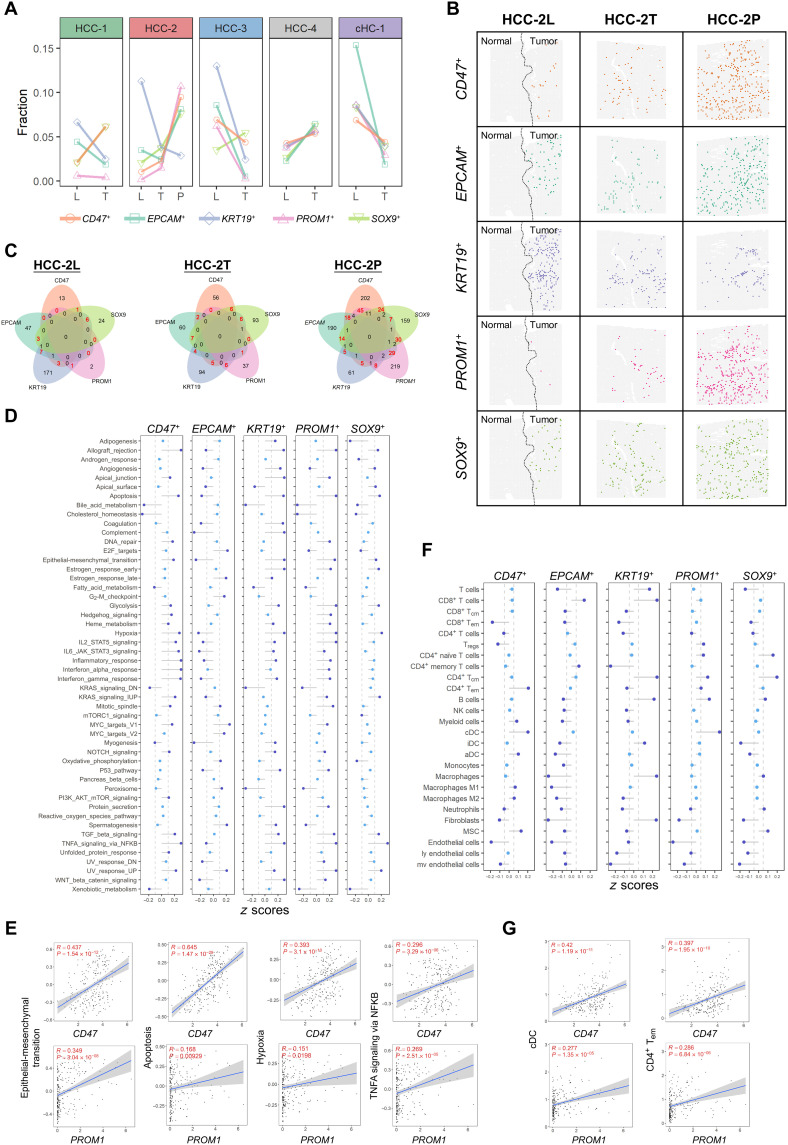
Fig. 5. Functional analysis of heterogeneous CSC niches in PLCs.
(A) Fractions of the positive spots of the five CSC markers in indicated sections. (B) Distribution of the putative CSC niches (spots) of five CSC markers in the L/T/P sections of HCC-2. (C) Venn diagrams of the positive niches of the five CSC markers in L/T/P sections of HCC-2. The numbers of double-positive spots were highlighted in red. (D) Hallmark pathway activities (average z scores of HCC-2P) in the five types of CSC niches. TGF, transforming growth factor; TNFA, tumor necrosis factor a; NFKB, nuclear factor kappa B subunit; UV, ultraviolet; IL6, interleukin 6; STAT3, signal transducer and activator of transcription 3; PI3K, phosphatidylinositol 3-kinase; mTOR, mammalian target of rapamycin. (E) Scatter plots showing the correlation between hallmark pathway activities and the expression levels of the CSC markers (CD47 and PROM1) on bulk sequencing data of 239 HCC cases. Linear regression and Pearson correlation were used to measure their relation. (F) Cell type enrichment scores (average z scores of HCC-2P) in the five types of CSC niches. MSC, mesenchymal stem cells. (G) Scatter plots showing the correlation between cell type enrichment scores and the expression levels of the CSC markers (CD47 and PROM1) on bulk sequencing data of 239 HCC cases. Linear regression and Pearson correlation were used to measure their relation.