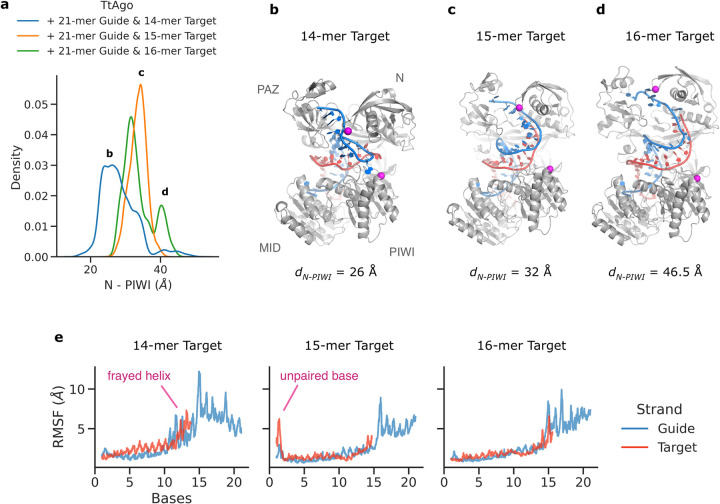
Fig 5. H-REMD simulations of ternary complexes with 14, 15 or 16 bp between guide and target DNA strands.
(a) Sampled distribution of the N-PIWI domain distances in the three ternary complexes Argonaute with varying target length. The distances are measured between α-carbon atoms of the two residues M82 from N and D552 from PIWI domain, indicated as magenta spheres (in panels b-d). The distance distributions indicate on average a shift towards N-PIWI opening as the target-guide base pairing is increased to 15 or 16 bp. (b-d) This is also evident in snapshots taken from the simulations of 14, 15, and 16-mer target DNA ternary complexes. The guide and target strands are shown in blue and red cartoons). (e) Root mean square fluctuations (RMSF) of each base for all the DNA strands during the H-REMD (in the reference replica). The RMSF values were calculated with respect to the average structure.