Abstract
Background
The gut microbiota constitutes a dynamic microbial system constantly challenged by environmental conditions, including physical exercise. Limited human studies suggest that exercise could play a beneficial role for gut health, increasing microbial diversity, even if the effects of exercise on gut microbial microorganisms depends on its intensity and duration. This study aimed to investigate the effects of nine weeks of high-intensity interval exercise on gut microbiota composition in healthy young adults.
Methods
The gut microbiota composition of seventeen healthy male college students was analysed before and after nine weeks of high-intensity interval cycling training by 16S rRNA amplicon sequencing. PERMANOVA for repeated measures was used to test pre-post differences in the relative abundance of all taxonomic levels, and correlations between variations in microbial composition and physical and dietary features were also assessed.
Results
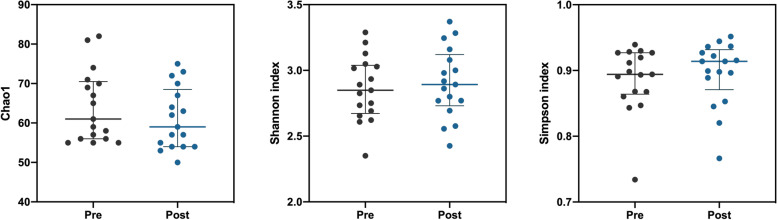
Physical exercise induced changes in microbiota composition, at all taxonomic levels analysed (phyla: F [1, 32]=3.97, p=0.029; classes: F [1, 32]=3.39, p=0.033, orders: F [1, 32]=3.17, p=0.044, families: F [1, 32]=1.54, p=0.037, genera: F [1, 32]=1.46, p=0.015, species: F [1, 32]=1.38, p=0.007). Conversely, no differences were found between pre and post-training conditions for microbial community richness (Chao1: V=105, p=0.06) or diversity (Shannon index: V=62, p=0.52; Simpson index: V=59, p=0.43). Changes in the relative abundance of eighteen genera were correlated to changes of twenty environmental factors grouped in physical features, sport-related features, and dietary features.
Conclusions
Nine weeks of high-intensity exercise induced modifications in gut microbiota composition in healthy male college students, shifting the gut microbial population towards a healthier microbiome with benefit to human health in general.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12970-021-00471-z.
Keywords: gut microbiota, physical exercise, diet habits, high-intensity interval exercise
Background
The human gut microbiota is composed of over a hundred trillion microbial cells, and its association with human health has been increasingly studied in the last decades [1, 2]. The gut microbial community significantly influences the host's metabolism and immune system, representing a risk or protective factor to several immune-allergic and metabolic disturbances [3, 4]. Its composition is highly subjective, and it is associated with environmental and behavioural factors, such as age, diet [1], supplementation [5] and physical exercise [6]. The latter was linked with positive changes in the gut microbiota diversity and community, both in animal and human studies [7, 8]. Among humans, positive correlations were reported between bacterial diversity, butyrate-producing bacteria and cardiorespiratory fitness (VO2max) [9], and higher turnover of carbohydrates and proteins, and concentrations of short-chain fatty acids in athletes compared to sedentary controls [10]. An observational study conducted on elite male rugby players showed that athletes have a higher abundance of Firmicutes and lower levels of Bacteroidetes than non-athletes healthy controls [11]. In a recent study, higher microbial diversity, different taxonomic and functional composition, and a strong association with athletes performance in elite athletes with respect to non-elite was reported when faecal samples of nineteen individuals pertaining to three cohorts (adult elite, youth elite and non-elite athletes) were compared [1]. A review published by Mitchell and colleagues in 2019 [12] reported only one longitudinal intervention study [13] where dietary intake and exercise were monitored, and participants tested pre and post-training. Indeed, an increase in beta-diversity after six weeks of endurance training in obese individuals was observed [13]. Authors controlled the dietary intake for three days before the stool collection to minimise the likely confound of differences in dietary intake on microbial composition; nevertheless, diet monitoring for a more extended period, before and during the training, was missing. This point should be crucial in these intervention studies, as changes in microbiota composition could not be attributed exclusively to the exercise but also to changes in the dietary intake, which was shown to be influenced by exercise itself [14]. Other studies were recently published investigating the effect of light-to-moderate intensity exercise training on changes in microbiota profile in overweight adults [15], women [16], and obese children [17]. Rettedal et al. [18] investigated the effect of a short-term high-intensity interval training period on the gut bacterial composition of lean and overweight men, failing to find changes neither in diversity nor composition of the microbiota. Considering the above-presented studies, a systematic review recently published [19] have pointed out the need for longer duration and higher intensity training studies to analyse significant taxonomic changes in microbiota composition further.
Hence, this study aimed to investigate the effects of 9-week high-intensity interval exercise on gut microbiota composition in active, healthy young adults. To the authors' knowledge, this is the first training study with a structured exercise period and daily monitoring of training and dietary intake.
Methods
Participants
Eighteen healthy college students (22±2 years, 175.7±6.7 cm, 69.1±10.1 kg, 22.3±2.7 kg/m2) were recruited. The inclusion criterion was to be sedentary: by completing a specific survey, subjects declared not performing more than one 60 min low-intensity training session per week in the three months before the start of the study. Baseline VO2max values confirmed the low training level of the participants. Exclusion criteria were musculoskeletal injuries, major cardiovascular diseases, upper respiratory infections, smoking in the past three months. The participants were advised to maintain their dietary routine and to refrain from other sports activities except the sessions scheduled for the experimental design. After a medical health screening, all participants signed a written informed consent to take part in the study, which was approved by the Ethics Committee of the University of Urbino Carlo Bo, Italy (no. 02/2017, approved on July 10, 2017) and was conducted in accordance with the Declaration of Helsinki for research with human volunteers (1975).
Study design
This was a longitudinal training study in which gut microbiota was analysed before and after nine weeks of high-intensity interval cycling training. Before and after the training period, body composition and maximal oxygen consumption (VO2max) were assessed. Participants arrived at the laboratory in the morning, after six or more hours of fasting. Body composition was measured through electrical bioimpedance (BIA101 Sport Edition, Arkray, Kyoto, Japan). They were asked to drink enough fluids over the 24h prior to the measurement to ensure a normal hydration state, and to abstain from alcohol and caffeine in the 8 hours before the test. After completing the body composition assessment, subjects received a standardised breakfast consisting of jam tart (~135 g per serving) and 400 ml of fruit juice. Two hours later, they started the testing session. Each participant performed a maximal graded exercise test to assess VO2max, peak power output (Wpeak) and power at lactate thresholds (WLT1 and WLT2). The test was performed on an electronically-braked ergometer (SRM Italia, Lucca, Italy) at 75W, and power was increased by 25W every 3 min until volitional exhaustion. Oxygen consumption was monitored breath-by-breath using a wearable metabolic cart (Cosmed K4b2, COSMED, Rome, Italy), and blood lactate was measured in the last 15 seconds of each stage from the tip of the index finger using a blood lactate meter (Lactate-Pro, Arkray, Kyoto, Japan). For more detailed information, see Gervasi et al. [20].
Training characteristics
Thirty-six indoor cycling training sessions during nine weeks were performed. The training program consisted of three mesocycles (three weeks each), on which both frequency and duration of the sessions increased: 3 × 55 min sessions per week in the first mesocycle, 4 × 60 min sessions per week in the second mesocycle, 5 × 70 min sessions per week in the last mesocycle. Each session was structured with a warm-up, a high-intensity interval exercise and a cool down. Intervals were performed at different intensities and durations to achieve 20% of the total training volume at an intensity above the second lactate threshold (LT2). Heart rate (HR) was recorded during each session, and HR values were used by the participants to match the requested intensity on each interval, according to their individual training zones defined with the incremental test.
Dietary monitoring
Diet was daily monitored from two weeks before the beginning of the training period until the end of the study, and it was performed by call interviews each day after dinner. Information about food and drink consumed were collected and then processed through MètaDieta software (METEDA Srl, San Benedetto del Tronto, Italy), and the following variables were considered for the analyses: total energy intake, macronutrients quantity, starch, soluble and insoluble fibres, saturated, monounsaturated, polyunsaturated fat, Omega-3, Omega-6, iron, vitamin A, C, D, E and amino acids (alanine, valine, isoleucine, leucine, tryptophan).
Faeces sample collection and DNA extraction
Faeces samples were collected at baseline and after the nine weeks of training. Faecal samples were collected in a sterilized 50 mL tube without additives, ethanol or stabilizing solutions, immediately stored and placed on frozen packages and delivered to the laboratory to be stored at -80 °C within two hours. For 16SrDNA sequencing, the total microbial DNA of the samples was extracted using the QIAamp PowerFecal DNA Kit (Qiagen) following the manufacturer’s protocol. After assessing DNA concentration and purity, samples were stored at -80° until processing.
16S rRNA gene sequence data processing
The V3–V4 hypervariable regions of the bacterial 16S ribosomal RNA gene were amplified for each DNA sample using the universal primers, as reported in Klindworth et al. [21]. Agencourt AMPure XP beads (Beckman Coulter, Milan, Italy) were used to purify PCR amplicons. The latter was used for a second PCR to barcode the libraries using the Illumina dual-index system (Nextera XT Index Kit, Illumina Inc., San Diego, CA, USA) necessary for multiplexing. The purified DNA products were then subjected to a further PCR to attach dual Illumina indices (Nextera XT Index Kit, Illumina Inc., San Diego, CA, USA) necessary for multiplexing. According to the manufacturer's instructions, paired-end sequencing (2 × 300 cycles) was carried out on an Illumina MiSeq instrument (Illumina Inc.). Sequences were demultiplexed based on index sequences, and FASTQ files were generated. Sequences were then analysed using QIIME 1.9.1 software and following the pipeline [22]. Sequences were quality filtered to remove short and long sequences (truncated sequence should be > 75 bases long), uncorrected barcodes, ambiguous bases, and sequences with primer mismatches. Chimeric sequences were identified and removed from all downstream analyses. To that end, both de novo and reference chimaeras were identified and removed employing usearch 6.1 and the GreenGenes database (v. 105 13-8, 97% identity). The latter was also used for open reference OTU (Operational Taxonomic Units) picking and taxonomic assignment through UCLUST.
Statistical analyses
One subject was excluded from the analyses because of faecal sample collection errors. Alpha diversity indexes (Chao1, Shannon H and Simpson) were calculated using the diversity function of the vegan R package [23], both in pre and post-training conditions. Differences among the pre and post-training diversity indexes were tested using a Wilcoxon signed-rank test for paired data. Bray-Curtis dissimilarity was calculated using vegdist R function; Permutational Analysis of Variance (PERMANOVA) for repeated measures was used to test pre-post differences in the relative abundance of all taxonomic levels (phyla, classes, orders, families, genera and species), using adonis R function with subjects’ ID as strata for pairing. Post-hoc comparisons at phylum and genus levels were performed using a Wilcoxon test, and effect sizes were calculated for all comparisons using the wilcox_effsize function of rstatix [24] and coin R packages [25]. Common interpretations of Wilcoxon effect sizes (r) are: 0.10 - 0.3 (small effect), 0.30 - 0.5 (moderate effect) and >= 0.5 (large effect) [26]. A ternary plot was built to visualise the co-variations of the relative abundances in the three most dominant phyla, using the ggtern R package [27]. Pre-post comparisons between all phyla present in our sample were graphically reported as box plots, using the ggplot2 R package [28]. Genera were then filtered for a relative abundance higher or equal to 1%, in pre or post-training conditions, for the subsequent analyses. A correlation matrix was calculated on the post-pre differences of the relative abundance of the 29 remaining genera, and it was graphically represented on a non-parametric correlation plot, all using the corrplot R package [29]. To assess post-pre changes in global genera relative abundances, a non-metric dimensional scaling (nMDS) was performed, with the Bray-Curtis distance, using the metaMDS function of the vegan R package [23]. Finally, from an exploratory point of view, to assess correlations between variations in microbial composition and physical and dietary features, a circular correlation plot was built, using the chordDiagram function of the circlize R package [30]. Only significant correlations (Spearman r ≥ 0.485, for n=17; p≤0.05, two-tailed) are reported in the circle plot for visual clarity. Statistical analyses were performed using R Studio 1.4; the significance threshold was fixed at the standard level of 0.05.
Results
Exercise shapes gut microbiota composition without affecting richness and diversity indices
Seventeen healthy male subjects completed thirty-six indoor cycling training sessions during nine weeks. Training sessions were: 3 × 53.1±1.3min sessions per week for the first three weeks, 4 × 59.1±1.2min sessions per week for the second three weeks, 5 × 68.2±1.4min sessions per week for the last three weeks.
By sequencing the V3-V4 region of the 16S rRNA gene of Archaea and Bacteria from 34 faecal samples collected from 17 participants, we identified 11 phyla, 22 classes, 34 orders, 62 families and 123 genera. Physical exercise induced changes in microbiota composition, at all taxonomic levels analysed (PERMANOVA for paired data, Bray-Curtis dissimilarity index; phyla: F (1, 32)=3.97, p=0.029; classes: F (1, 32)=3.39, p=0.033, orders: F (1, 32)=3.17, p=0.044, families: F (1, 32)=1.54, p=0.037, genera: F (1, 32)=1.46, p=0.015, species: F (1, 32)=1.38, p=0.007). Conversely, no differences were found between pre and post-training conditions for microbial community richness (Chao1: Wilcoxon test, V=105, p=0.06) or diversity (Shannon index: V=62, p=0.52; Simpson index: V=59, p=0.43). Data are presented in Fig. 1.
Fig. 1.
Comparisons of Chao1 index (left), Shannon index (middle) and Simpson index (right) of microbial communities in pre and post-training conditions
Phylum level
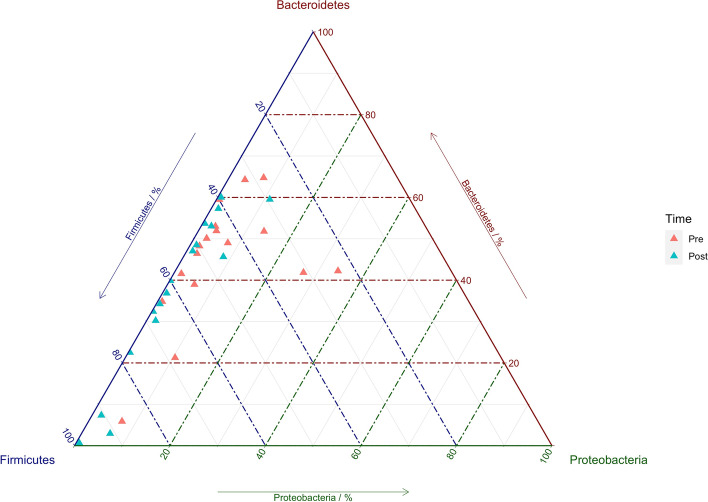
The most abundant bacterial phyla identified were constituted by Firmicutes, Bacteroidetes, Proteobacteria: average relative abundances were 45.5% and 56.0% for Firmicutes, 43.5% and 35.3% for Bacteroidetes, and 7.6 and 2.2% for Proteobacteria, respectively in the pre and post-training conditions. The distribution of samples in the two conditions, based on the relative abundance of the three most dominant phyla, is represented in the ternary plot below (Fig. 2).
Fig. 2.
Ternary plot showing the distribution of samples in pre and post-training conditions, based on the relative abundance of three dominant phyla
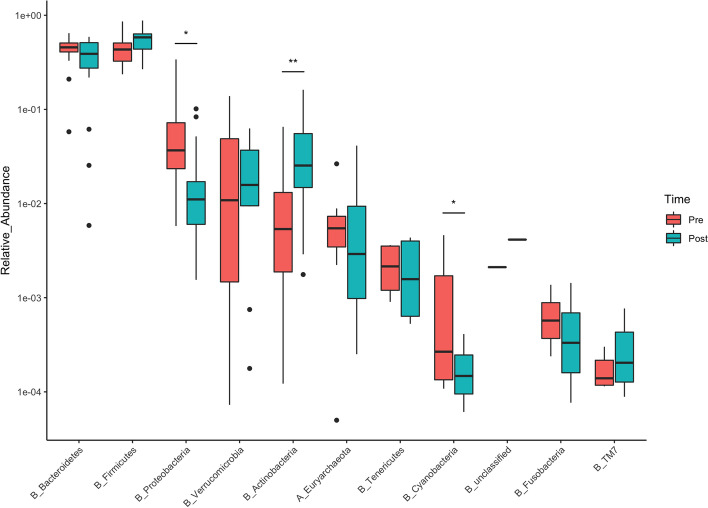
After checking for overall significance with the PERMANOVA test (see point 3.2), post-hoc univariate comparisons have been performed for all phyla with a Wilcoxon signed-rank test for paired data. Significant differences in relative abundances between post- and pre-training conditions were found for Actinobacteria (+3.38%; p=0.003, ES=0.72 (large effect)), Cyanobacteria (-0.05%; p=0.025, ES=0.54 (large effect)) and Proteobacteria (-5.45%; p=0.01, ES=0.614 (large effect)). Despite not reaching statistical significance, moderate effect sizes were found for Bacteroidetes (-8.28%; p=0.05; ES=0.47 (moderate effect)), and Firmicutes (+10.46%; p=0.08; ES=0.43 (moderate effect)). These results should be read keeping in mind the low sample size, which influences the significance of the tests. All the relative abundances of the phyla in pre and post-training conditions are reported in the box plots below (Fig. 3).
Fig. 3.
Box plots of relative abundances of phyla (expressed in logarithmic scale for visual clarity). Stars above the boxes represent statistical significance (*p<0.05, **p<0.01)
The Firmicutes:Bacteroidetes (F/B) ratio was calculated and compared in pre and post-training conditions. Median F/B ratio in pre-training was 0.89 (Q1: 0.67, Q3: 1.88), and in post-training was 1.50 (Q1: 0.85, Q3: 2.24) (Wilcoxon test, V=120, p=0.039, ES=0.50 (moderate effect)).
Genus level
As for phyla above, pre vs post-training comparisons were performed with a Wilcoxon test for paired data. A cut-off on the mean relative abundance of each genus was used, filtering data for an abundance equal or higher than 1%, in pre or post-training conditions. Twentynine genera were selected based on this criterion for further analyses. Bifidobacterium, Parabacteroides, Prevotella, Dorea, Lachnospira, unclassified Ruminococcaceae, Phascolarctobacterium and Sutterella were significantly different in post- vs pre-training. Descriptive (median (Q1 - Q3)) and test statistics for all the 29 genera are reported in Table 1. A list of all 123 genera comparisons is reported as an Additional file (Additional file 1).
Table 1.
Changes in relative abundances of more represented genera (filtered for mean relative abundance >1% in pre or post-training condition). Differences between pre and post-training were tested using Wilcoxon paired tests. Effect sizes are also reported, with 95% confidence intervals
| Pre | Post | p | Effect Size (95% CI) | |
|---|---|---|---|---|
| Actinobacteria | ||||
| g_Bifidobacterium | 0.37 (0.16 - 1.27) | 2.33 (1.41 - 4.91) | 0.004 | 0.71 (0.37 - 0.88) |
| Bacteroidetes | ||||
| g_Bacteroides | 32.3 (20.1 - 38.4) | 21.1 (8.5 - 34.2) | 0.163 | 0.34 (0.03 - 0.71) |
| g_Parabacteroides | 2.43 (1.04 - 4.01) | 1.39 (0.55 - 2.39) | 0.019 | 0.57 (0.15 - 0.83) |
| g_Prevotella | 0.42 (0.01 - 4.77) | 0.03 (0.01 - 2.45) | 0.041 | 0.48 (0.15 - 0.82) |
| f_Rikenellaceae; g_ | 2.86 (0.82 - 4.19) | 3.18 (1.27 - 4.17) | 0.523 | 0.15 (0.02 - 0.54) |
| Firmicutes | ||||
| g_Lactobacillus | 0.00 (0.00 - 0.01) | 0.00 (0.00 - 0.01) | 0.866 | 0.05 (0.01 - 0.55) |
| g_Streptococcus | 0.14 (0.07 - 0.48) | 0.23 (0.14 - 1.01) | 0.435 | 0.19 (0.01 - 0.67) |
| g_Turicibacter | 0.18 (0.05 - 0.51) | 0.19 (0.04 - 0.62) | 0.586 | 0.13 (0.01 - 0.59) |
| f_Clostridiaceae; g_ | 0.38 (0.07 - 1.25) | 0.27 (0.07 - 0.81) | 0.723 | 0.09 (0.01 - 0.55) |
| f_Lachnospiraceae; g_[Ruminococcus] | 0.88 (0.75 - 2.02) | 1.07 (0.85 - 1.84) | 0.381 | 0.21 (0.01 - 0.61) |
| g_Blautia | 2.54 (1.78 - 3.14) | 3.88 (1.83 - 9.50) | 0.062 | 0.45 (0.07 - 0.77) |
| g_Coprococcus | 2.15 (1.29 - 3.16) | 2.73 (1.65 - 3.52) | 0.113 | 0.38 (0.02 - 0.75) |
| g_Dorea | 0.38 (0.16 - 0.62) | 0.67 (0.37 - 1.82) | 0.019 | 0.57 (0.06 - 0.88) |
| g_Lachnospira | 0.84 (0.15 - 2.36) | 0.41 (0.03 - 0.50) | 0.007 | 0.65 (0.31 - 0.87) |
| g_Roseburia | 2.70 (1.39 - 6.02) | 1.81 (1.12 - 4.21) | 0.227 | 0.29 (0.02 - 0.64) |
| f_Lachnospiraceae; g_ | 3.25 (1.09 - 4.17) | 2.79 (2.28 - 4.64) | 0.831 | 0.05 (0.00 - 0.51) |
| f_Peptostreptococcaceae; g_ | 0.97 (0.38 - 1.55) | 1.08 (0.46 - 1.86) | 0.593 | 0.17 (0.02 - 0.72) |
| g_Faecalibacterium | 3.25 (2.54 - 4.21) | 3.76 (2.99 - 6.41) | 0.266 | 0.27 (0.02 - 0.65) |
| g_Oscillospira | 1.73 (0.91 - 2.43) | 2.08 (0.45 - 4.49) | 0.246 | 0.28 (0.04 - 0.76) |
| g_Ruminococcus | 0.91 (0.26 - 2.59) | 1.73 (0.68 - 4.32) | 0.055 | 0.46 (0.08 - 0.86) |
| f_Ruminococcaceae; g_ | 3.27 (1.98 - 6.06) | 7.48 (4.73 - 8.36) | 0.003 | 0.72 (0.36 - 0.88) |
| o_Clostridiales; f_; g_ | 0.83 (0.40 - 2.88) | 0.60 (0.34 - 2.29) | 0.493 | 0.17 (0.01 - 0.62) |
| g_Dialister | 0.17 (0.01 - 3.17) | 2.77 (0.00 - 3.46) | 0.972 | 0.03 (0.00 - 0.51) |
| g_Phascolarctobacterium | 0.37 (0.00 - 2.21) | 0.03 (0.00 - 0.58) | 0.039 | 0.55 (0.13 - 0.85) |
| g_Veillonella | 0.03 (0.01 - 0.16) | 0.00 (0.00 - 0.03) | 0.084 | 0.39 (0.04 - 0.83) |
| f_Erysipelotrichaceae; g_ | 0.29 (0.12 - 0.77) | 0.60 (0.23 - 1.15) | 0.068 | 0.44 (0.05 - 0.78) |
| Proteobacteria | ||||
| g_Escherichia | 0.46 (0.15 - 3.68) | 0.13 (0.02 - 0.66) | 0.062 | 0.45 (0.03 - 0.77) |
| g_Sutterella | 0.71 (0.22 - 1.45) | 0.15 (0.12 - 0.38) | 0.039 | 0.50 (0.12 - 0.87) |
| Verrucomicrobia | ||||
| g_Akkermansia | 0.00 (0.00 - 0.42) | 0.07 (0.00 - 1.77) | 0.767 | 0.08 (0.01 - 0.58) |
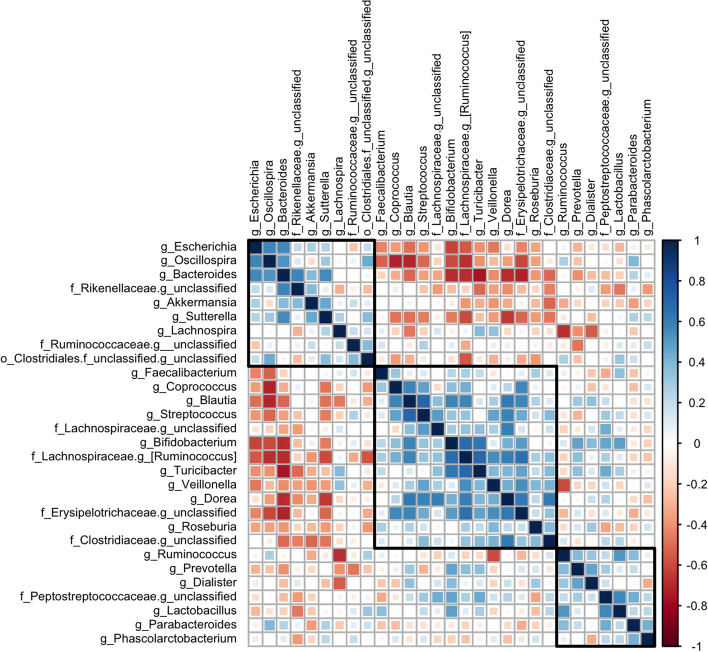
Non-parametric Spearman correlations of delta changes (post-pre) in the relative abundances of the 29 genera have been calculated to highlight subgroups of genera showing a similar change direction; correlations are presented in Fig. 4. Three clusters of genera can be identified: a first one (top left) is mainly composed of Bacteroides, Lachnospira, Oscillospira, Sutterella, Escherichia and Akkermansia; a second one (centre) in which the genera belonging to Firmicutes phylum (Streptococcus, Turicibacter, Blautia, Coprococcus, Dorea, Roseburia, Faecalibacterium, Veillonella) are prevalent, with the addition of Bifidobacterium (of the Actinobacteria phylum); finally, a third group (bottom right) formed by Parabacteroides, Prevotella, Lactobacillus, Ruminococcus, Dialister and Phascolarctobacterium can be highlighted. Interestingly, the genera of the first and second clusters are negatively correlated, while almost no correlation can be identified between the genera of the third cluster and the other two. A closer look at the clusters shows that most of the genera of the first cluster are reduced in abundance in post-training (except for f_Ruminococcaceae.g_unclassified), while the genera of cluster two show an opposite trend (except for Roseburia and Veillonella).
Fig. 4.
Correlation plots of delta (post-pre) at genus level: genera filtered for relative abundances over 1% in pre or post-training condition. Genera are ordered with hierarchical clustering (hclust method), and three subgroups of genera are highlighted
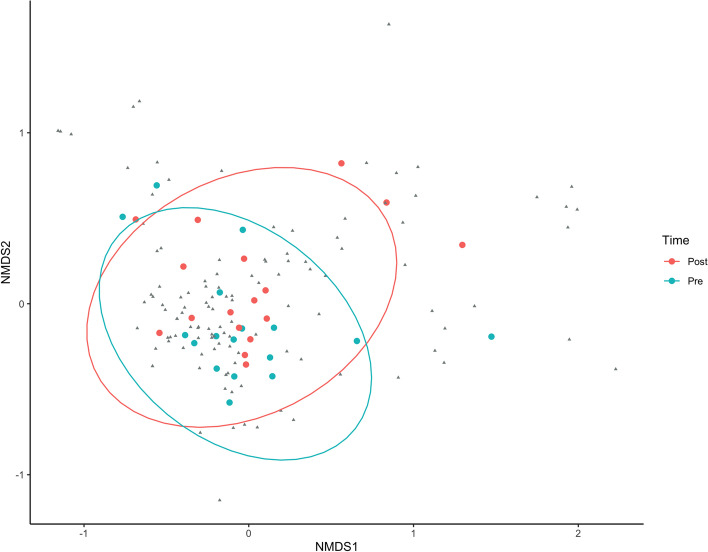
Non-metric multidimensional scaling (nMDS) is a method to explore and visualise dissimilarities of data matrix; it is a powerful tool to show specific patterns in multiple datasets. We used this approach to analyse the specific gut microbiota distribution patterns in pre-post training conditions. In Figure 5, a high dimensional space has been reduced into 2-dimensional space without too much loss of information (stress = 0.133, stress plot R2 = 0.98). Each bacterial genus was reported as small grey points, while subjects, in pre and post measurements, were reported in blue and red points in pre and post measurements, respectively. The ellipses referring to the pre-post conditions showed how some subjects had changed their position, while the majority fall into the common area of the two ellipses: From this, it can be inferred that, although PERMANOVA analysis showed a significant global variation (p<0.015), the training effect was noticeable only in a subgroup of subjects.
Fig. 5.
Non-metric dimensional scaling (PCoA) of Bray Curtis distance at the genus level, visualising the microbial composition in the pre- (light blue) and post-training (red) conditions
Associations among changes in microbiota composition, dietary features and performance
The training period induced modifications in the subjects' physical composition, dietary habits, and performance indexes (Table 2). Even though assessing changes in these parameters was not among the aims of this study, these results must be described to allow a more straightforward understanding of the following associations among these and the changes in microbial composition. Nine weeks of high-intensity training induces a reduction of fat mass (-19.4%) and a gain of muscular mass (+3.7%), and an increase in relative peak power output (Wpeak/kg; +20.5%), and power at first (+45.2%) and second (+37.5%) lactate thresholds. In addition, as previously in a previous publication from our group (see Donati Zeppa et al. [14]), the training period promoted a spontaneous modulation of food choices. A significant increase in protein (+15.8%) and carbohydrate (+23.2%) intake has been observed during the training period. Energy and fats showed a similar trend, without a significant increase. A significant augment of fibres and vitamin C has been observed, suggesting higher consumption of fruit and vegetables during the training with respect to the pre-training.
Table 2.
Environmental factors in pre and post-training. Mean ± SD, Δ% and t-value (Cohen's D Effect Size) are reported
| Variable | Pre-training | Post-training | Δ% | t-value (ES) |
|---|---|---|---|---|
| Physical features | ||||
| Weight (kg) | 68.1 ± 9.5 | 67.4 ± 7.5 | - 0.6% | -1.182 (0.287) |
| Body Mass Index (kg/m2) | 22.1 ± 2.6 | 22.1 ± 2.2 | 0.2% | -0.059 (0.014) |
| Fat Mass (%) | 15.8 ± 5.4 | 12.5 ± 4.6 | -19.4% | -4.170 (1.011) |
| Muscular Mass (kg) | 42.1 ± 4.9 | 43.4 ± 3.8 | 3.7% | 2.708 (0.657) |
| Sport-related features | ||||
| VO2max (ml/min) | 2965 ± 418 | 3133 ± 344 | 7.0% | 1.935 (0.469) |
| VO2max (ml/kg/min) | 44.2 ± 8.0 | 46.5 ± 5.8 | 6.9% | 1.723 (0.418) |
| Wpeak (W/kg) | 3.4 ± 0.4 | 4.1 ± 0.5 | 20.5% | 11.147 (2.703) |
| LT1 (W/kg) | 1.2 ± 0.3 | 1.7 ± 0.4 | 45.2% | 3.997 (0.969) |
| LT2 (W/kg) | 2.0 ± 0.5 | 2.7 ± 0.4 | 37.5% | 4.860 (1.179) |
| Dietary features | ||||
| Energy (kcal) | 2098 ± 481 | 2413 ± 790 | 16.2% | 2.048 (0.497) |
| Protein (g) | 84.4 ± 17.2 | 98.2 ± 30.0 | 15.8% | 2.642 (0.641) |
| Carbohydrate (g) | 266.9 ± 60.7 | 322.2 ± 100.9 | 23.2% | 2.548 (0.618) |
| Starch (g) | 129.7 ± 47.1 | 158.3 ± 61.9 | 40.5% | 1.967 (0.477) |
| Fat (g) | 78.9 ± 24.4 | 84.8 ± 36.3 | 9.4% | 0.833 (0.202) |
| Saturated Fats (g) | 23.0 ± 7.6 | 24.6 ± 10.2 | 10.1% | 0.767 (0.186) |
| Monounsaturated Fats (g) | 27.0 ± 9.3 | 28.5 ± 13.5 | 11.5% | 0.517 (0.126) |
| Polyunsaturated Fats (g) | 8.5 ± 2.9 | 10.0 ± 4.5 | 28.3% | 1.152 (0.279) |
| Omega-3 (% kcal/kcal tot) | 0.5 ± 0.2 | 0.4 ± 0.1 | 4.2% | -0.987 (0.239) |
| Omega-6 (% kcal/kcal tot) | 3.1 ± 0.9 | 3.0 ± 0.5 | 5.8% | -0.201 (0.049) |
| Fibres (g) | 8.4 ± 4.4 | 11.7 ± 8.4 | 48.4% | 2.203 (0.534) |
| Iron (mg) | 8.4 ± 2.6 | 10.0 ± 4.1 | 26.6% | 1.982 (0.481) |
| Vitamin A (mcg) | 772 ± 459 | 920 ± 540 | 44.1% | 1.345 (0.326) |
| Vitamin C (mg) | 48.7 ± 24.0 | 84.5 ± 51.7 | 117.4% | 3.150 (0.764) |
| Vitamin D (mg) | 1.7 ± 1.4 | 1.8 ± 0.7 | 54.9% | 0.535 (0.130) |
| Vitamin E (mg) | 7.0 ± 3.0 | 8.7 ± 4.9 | 29.7% | 1.845 (0.448) |
| Aminoacids | 12509 ± 4183 | 15121 ± 4916 | 28.6% | 2.641 (0.641) |
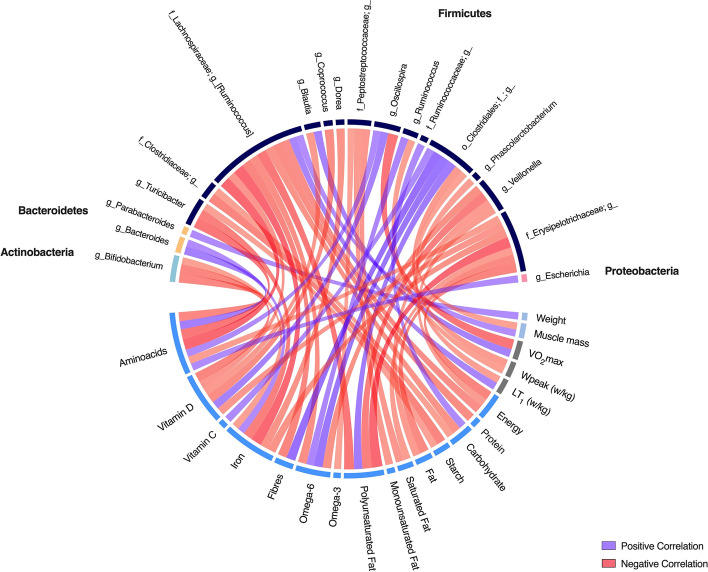
The correlations among the post-pre changes in the twenty-six factors presented in Table 2 and the bacterial taxonomic composition at the genus level (using the same cut-off of 1% in relative abundance as above) were investigated. Twenty factors and eighteen genera left after removing non-significant correlations (Spearman r ≥ 0.485, for n=17; p≤0.05, two-tailed), and the remaining are graphically presented as a circus plot in Fig. 6.
Fig. 6.
Spearman correlations between post-pre training delta of environmental factors and microbiota (genus level). Twenty-nine genera were selected for relative abundance >1%. 26 environmental factors were used for the correlation analysis, grouped in physical features [4], sport-related features [5], and dietary features [17]. Only significant correlations are reported in the figure. Sectors have different colours for groups of environmental factors and different phyla for simplicity. Purple links represent positive correlations, and red links represent negative correlations.
Among the physical features, a slight body mass reduction was accompanied by a decrease in abundance of Parabacteroides (r=0.50), while the muscle mass gain was associated with an increase of [Ruminococcus] (r=0.49) and a reduction of an unclassified genus of the Clostridiales order (r=-0.49). Among the indexes of performance, improvements in VO2max and power at LT1 were positively associated with increased Blautia (r=0.55) and [Ruminococcus] (r=0.53), respectively. In contrast, VO2max and LT1 were negatively correlated to Oscillospira (r=-0.69) and Phascolarctobacterium (r=-0.50), respectively. The increase in peak power output was negatively correlated with Ruminococcus (r=-0.50) and an unclassified genus of the Clostridiales order (r=-0.54). In addition, considering absolute values, without taking into consideration post-pre differences, Bifidobacterium was positively correlated with peak power output (r=0.34), while Parabacteroides was negatively correlated with peak power output (r=-0.38) and VO2max (r=-0.37).
Among the dietary features, the increase in energy intake was negatively correlated with [Ruminococcus] (r=-0.59), Veillonella (r=-0.53) and an unclassified genus of the Erysipelotrichaceae family (r=-0.63). In addition, unclassified Ruminococcus showed negative correlations also with protein (r=-0.60), carbohydrate (r=-0.57), polyunsaturated fats (r=-0.69), omega-6 (r=-0.59), fibres (r=-0.63), iron (r=-0.72), vitamin D (r=-0.55), and amino acids (r=-0.74). Carbohydrate intake was also positively associated with an unclassified genus of the Ruminococcaceae family (r=0.51) and negatively correlated with an unclassified genus of the Clostridiaceae family (r=-0.50). All the correlations are reported as an Additional file (Additional file 2).
Discussion
This study aimed to investigate changes in gut microbiota profiles, richness and diversity indices following nine weeks of training intervention and draw associations between microbial modifications, dietary habits, and sports performance changes. To our knowledge, this is the first training study that explored the effect of nine-week of high-intensity indoor cycling training on the microbiota composition in non-athlete healthy male college students, with a structured exercise period and daily monitoring of training and dietary intake. The results obtained suggest that this structured training induces partial changes to the relative abundance and community structure of gut microbiota, although no significant changes in α-diversity were found. The latter is in accordance with the results of Rettedal et al. [18], which to the authors' knowledge, is currently the only study in which the effects of a high-intensity training program on human gut microbiota were investigated. Indeed, the authors did not find any difference in α- or β-diversity after three weeks of HIIT in lean and overweight men. Other conflicting findings were reported by Yuan et al. [31] and Wang et al. [32], which found a lower and higher α-diversity (Simpson and Shannon indexes), respectively, after four and seven weeks of high-intensity exercise in mice. Greater microbiota α-diversity was reported in athletes [11], although no significant associations were reported between α-diversity and cardiorespiratory fitness levels [19].
Microbial composition
Differences between pre and post-training conditions were detected at the phylum level, with an increase of Actinobacteria and a reduction of Proteobacteria and Cyanobacteria. No significant changes were found for Firmicutes and Bacteroidetes, although upward and downwards trends can be observed, respectively. Despite the absence of significance at the univariate level, a significant increase in Firmicutes/Bacteroidetes ratio (F/B) was identified after the training period. This result is in agreement with other cross-sectional studies, which reported higher levels of Firmicutes and lower levels of Bacteroidetes (thus an increased F/B ratio) in professional rugby players with respect to high-BMI controls [11] and adult’s elite respect to young elite and non-elite rowers [1]. F/B ratio was reported to be positively correlated with a higher VO2max and with faecal total short-chain fatty acids [1]. An increase in the F/B ratio has also been observed by Huang et al. [33] in obese adolescents after six weeks of exercise intervention along with dietary restriction. Firmicutes are known to produce butyrate, considered a health-promoting molecule due to its role in regulating energy metabolism and increasing insulin sensitivity [34]. A decrease of Proteobacteria and Cyanobacteria and an increase of Actinobacteria have been observed after nine weeks of training in our study, suggesting the positive effect of exercise in promoting the host's health. Indeed, training promoted a decrease of the inflammatory process by reducing bacteria associated with pro-inflammation such as Proteobacteria and increasing bacteria involved in anti-inflammatory processes such as Actinobacteria [35]. In general, Actinobacteria - although they represent only a tiny percentage of the total microbial community - are key players in the maintenance of gut homeostasis, increasing tight junctions expression, regulating mucin biosynthesis and catabolism, providing energy for epithelial cells proliferation and stimulating the immune system [36]. Classes of this phylum, especially Bifidobacteria, are widely used as probiotics demonstrating beneficial effects in many pathological conditions, even if larger in vivo studies are needed to confirm such encouraging results. Moreover, Bifidobacteria show a non-negligible production of lactate, which can be metabolised by a group of bacteria defined as "lactate utiliser" to produce butyrate [36, 37].
At the genus level, to the authors' knowledge, to date, only one study investigated microbiota changes related to high-intensity interval training: indeed, Rettedal et al. [18] did not find any association between the top 50 most abundant genera and cardiorespiratory fitness markers, suggesting that a short-term (three weeks) high-intensity interval training do not impact the overall composition of the gut microbiome. In our study, a decrease in Parabacteroides has been correlated with weight reduction after the training period, a result that is in accordance with previous findings by Karvonen et al. [38]. Together with other markers, such as Bacteroides, Blautia, Alistipes, Romboutsia and Roseburia, Parabacteroides were reported to represent a common feature in obese patients with different metabolic disorders [39]. In contrast, it is worth mentioning that the training program in our study tended to increase Blautia, typically reduced in obese patients and related to a health microbiota profile. Notably, the increase of short-chain fatty acids producing bacteria, including Blautia and Allobaculum, has been reported to alleviate inflammation, insulin resistance, and obesity by reducing the intestinal endotoxins into the blood [17]. The training period also induced an increase in the abundance of the Dorea genus, which was previously reported to be negatively associated with insulin resistance [40]. Reduction in Prevotella found in our study is at odds with the results of Petersen et al. [41], who found that Prevotella abundance was positively correlated with the weekly training volume of competitive cyclists. However, it should be noted that in cyclists with a training volume less than ten h/week, Prevotella relative abundance was about 0.15%, a value comparable to the one we found in our sample. Furthermore, the Sutterella genus, previously identified as a common driver taxon in diabetes [42], showed a significant decrease after training. Lastly, the Akkermansia genus deserves mention, despite its slight increase after training did not result to be statistically significant. Indeed, Akkermansia was found to be one of the main exercise-responsive taxa in overweight women after six weeks of endurance training [16], and it has also been reported to be more abundant in male elite rugby players with respect to non-athletes [11].
Nutrition
Diet is a major factor influencing the gut microbiome and should be considered a confounding factor when interpreting the results of an exercise intervention on microbiota composition. Some changes in the gut microbiota may be due to differences in dietary intake, in addition to the exercise itself [43]. Moreover, it has previously shown that exercise, particularly high-intensity exercise, might promote a spontaneous change of dietary choices toward a healthier direction [14]. So, to isolate and understand the direction of these relationships is not a foregone conclusion. Some differences or changes in the gut microbiota that seem to be associated with exercise might therefore be due to differences or changes in dietary intake, especially plants and carbohydrates, rather than the exercise itself. A significant increase in protein (+15.8%), carbohydrate (+23.2%) fibres, and vitamin C intake has been observed during the training period, suggesting higher consumption of fruit and vegetables during the training with respect to the pre-training. The mutual interactions between exercise, gut microbiota and diet, are the core in optimising performance and health. In particular, the increase of fibre intake leads to a higher production of short-chain fatty acids (SCFA), butyrate, propionate and acetate, which mediates the metabolic cross-talk between the gut microbiota and skeletal muscle [44]. SCFA affects muscle function, exercise capacity, glycogen accretion in skeletal muscle and performance [45, 46]. Furthermore, SCFA has a positive effect on muscle anabolism. In particular, butyrate and acetate stimulate glucose uptake, increase lipolysis and insulin sensitivity [47]. The effect of SCFA on skeletal muscles is mediated by activating muscular AMP kinase and the deposition of proteins in skeletal muscle tissue [48]. Furthermore, SCFA production is also associated with decreased inflammation and gastrointestinal disorders [49, 50].
In our study, several macro and micronutrient changes in post-pre training were associated with specific gut microbiota genera. Carbohydrate intake increases post-training and positively correlates with Ruminococcaceae, a commensal butyrate-producing bacteria belonging to the Firmicutes phylum, that can ferment indigestible carbohydrates and are crucial for maintaining health. Bacteroides affects amino acids’ availability and profile by participating in their digestion and absorption and are positively correlated with amino acids intake. SCFA can also be produced from branched-chain amino acids (valine, leucine, and isoleucine); threonine renders propionate and butyrate, whereas glutamate, histidine, lysine, arginine, and alanine give rise to acetate and butyrate formation. The metabolic versatility of Bacteroides and the modulation of its metabolism through the adequate balance of dietary proteins and carbohydrates could impact human health [51]. However, due to the major differences in population characteristics, dietary features, and data collection procedures, comparisons with other studies in the literature are complex and – in a certain sense – misleading.
Performance indexes
The gut microbiome has been previously shown to be associated with fitness and performance measures, such as VO2max and VO2peak. For example, either Allen et al. [13] and Estaki et al. [9] reported positive correlations among VO2max, VO2peak and butyrate-producing bacteria. VO2max has also been correlated with a higher F/B ratio [52, 53], although studies showed contrasting results [54]. Our study did not find any correlation between VO2max and F/B ratio, but it should be considered that VO2max values in our sample were quite homogeneous. Conversely, a negative correlation between VO2max and Parabacteroides has been highlighted. Considering post-pre training changes, VO2max showed a positive correlation with Blautia and a negative correlation with Oscillospira. A positive correlation between Blautia and VO2peak was previously found by Yu et al. [54] in an elderly population. Relative power output at LT1 was instead correlated with f_Lachnospiraceae [Ruminococcus] and Phascolarctobacterium, positively and negatively, respectively. The former is a key butyrate-producing member of a family that has been shown to be more represented in aerobically fit individuals [9]. A study by Scheiman et al. reported a high abundance of Veillonella atypica in the post-exercise state in athletes after completing the Boston Marathon, suggesting a possible association with running performance [55]. In contrast, we did not find any change in Veillonella relative abundance after the training period. Although the mechanisms are still unknown, gut microbiota was suggested to have a bidirectional cross-talk with mitochondria during endurance exercise [45], and skeletal muscle bioenergetics might be modulated by exercise-induced alterations in microbiota composition [4, 56].
A limitation of this study is represented by the fact that it was not possible to deduce if microbial composition changes were related only to an exercise effect, to the changes in the dietary intake (possibly, exercise-driven) or a combination of both. A deeper understanding of this point should be of primary importance for future studies.
Conclusions
Physical activity modifies the gut microbiota, exerting health benefits on the host; however, the specific bacteria associated with exercise are not yet known. This work showed that nine weeks of high-intensity indoor cycling training induced modifications in gut microbiota composition in non-athlete healthy male college students. Some relevant bacterial taxa were modified after the training period shifting the gut microbial population towards a healthier microbiome. Understanding the direction in which microbes play a key role in influencing athletic performance is of particular interest to athletes who aim to improve their results in competition. However, such knowledge could be of benefit to human health in general.
Supplementary Information
Acknowledgements
Author Valerio Pazienza is supported by AIRC grant n. IG 23006 and by Italian Ministry of Health Ricerca Corrente program 2018-2020.
Authors' contributions
Conceptualization: SDZ and MG; Methodology: SA and DS; Formal analysis and investigation: DS, VP and MBLR; Writing - original draft preparation: SDZ and SA; Writing - review and editing: DA and VP; Supervision: GP, PG, MBLR, PS and VS. All authors read and approved the final manuscript.
Availability of data and materials
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Funding
No financial support was received for the conduct of this study or preparation of this article.
Declarations
Ethics approval and consent to participate
Ethics Committee of the University of Urbino Carlo Bo, Italy (no. 02/2017, approved on July 10, 2017). Signed informed consent was obtained from all subjects participating in the study.
Consent for publication
Not applicable.
Competing interests
None of the authors declare competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sabrina Donati Zeppa and Stefano Amatori contributed equally to this work.
References
- 1.Han M, Yang K, Yang P, Zhong C, Chen C, Wang S, et al. Stratification of athletes' gut microbiota: the multifaceted hubs associated with dietary factors, physical characteristics and performance. Gut Microbes. 2020;12(1):1–18. doi: 10.1080/19490976.2020.1842991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark A, Mach N. Exercise-induced stress behavior, gut-microbiota-brain axis and diet: a systematic review for athletes. J Int Soc Sports Nutr. 2016;13:43. doi: 10.1186/s12970-016-0155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donati Zeppa S, Agostini D, Gervasi M, Annibalini G, Amatori S, Ferrini F, et al. Mutual Interactions among Exercise, Sport Supplements and Microbiota. Nutrients. 2019;12(1). [DOI] [PMC free article] [PubMed]
- 6.Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms. 2019;7(1). [DOI] [PMC free article] [PubMed]
- 7.Mika A, Van Treuren W, Gonzalez A, Herrera JJ, Knight R, Fleshner M. Exercise is More Effective at Altering Gut Microbial Composition and Producing Stable Changes in Lean Mass in Juvenile versus Adult Male F344 Rats. PLoS One. 2015;10(5):e0125889. doi: 10.1371/journal.pone.0125889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clauss M, Gerard P, Mosca A, Leclerc M. Interplay Between Exercise and Gut Microbiome in the Context of Human Health and Performance. Front Nutr. 2021;8:637010. doi: 10.3389/fnut.2021.637010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estaki M, Pither J, Baumeister P, Little JP, Gill SK, Ghosh S, et al. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome. 2016;4(1):42. doi: 10.1186/s40168-016-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton W, Penney NC, Cronin O, Garcia-Perez I, Molloy MG, Holmes E, et al. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut. 2018;67(4):625–633. doi: 10.1136/gutjnl-2016-313627. [DOI] [PubMed] [Google Scholar]
- 11.Clarke SF, Murphy EF, O'Sullivan O, Lucey AJ, Humphreys M, Hogan A, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63(12):1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell CM, Davy BM, Hulver MW, Neilson AP, Bennett BJ, Davy KP. Does Exercise Alter Gut Microbial Composition? A Systematic Review. Med Sci Sports Exerc. 2019;51(1):160–167. doi: 10.1249/MSS.0000000000001760. [DOI] [PubMed] [Google Scholar]
- 13.Allen JM, Mailing LJ, Niemiro GM, Moore R, Cook MD, White BA, et al. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med Sci Sports Exerc. 2018;50(4):747–757. doi: 10.1249/MSS.0000000000001495. [DOI] [PubMed] [Google Scholar]
- 14.Donati Zeppa S, Sisti D, Amatori S, Gervasi M, Agostini D, Piccoli G, et al. High-intensity Interval Training Promotes the Shift to a Health-Supporting Dietary Pattern in Young Adults. Nutrients. 2020;12(3). [DOI] [PMC free article] [PubMed]
- 15.Cronin O, Barton W, Skuse P, Penney NC, Garcia-Perez I, Murphy EF, et al. A Prospective Metagenomic and Metabolomic Analysis of the Impact of Exercise and/or Whey Protein Supplementation on the Gut Microbiome of Sedentary Adults. mSystems. 2018;3(3). [DOI] [PMC free article] [PubMed]
- 16.Munukka E, Ahtiainen JP, Puigbo P, Jalkanen S, Pahkala K, Keskitalo A, et al. Six-Week Endurance Exercise Alters Gut Metagenome That Is not Reflected in Systemic Metabolism in Over-weight Women. Front Microbiol. 2018;9:2323. doi: 10.3389/fmicb.2018.02323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quiroga R, Nistal E, Estebanez B, Porras D, Juarez-Fernandez M, Martinez-Florez S, et al. Exercise training modulates the gut microbiota profile and impairs inflammatory signaling pathways in obese children. Exp Mol Med. 2020;52(7):1048–1061. doi: 10.1038/s12276-020-0459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rettedal EA, Cree JME, Adams SE, MacRae C, Skidmore PML, Cameron-Smith D, et al. Short-term high-intensity interval training exercise does not affect gut bacterial community diversity or composition of lean and overweight men. Exp Physiol. 2020;105(8):1268–1279. doi: 10.1113/EP088744. [DOI] [PubMed] [Google Scholar]
- 19.Aya V, Florez A, Perez L, Ramirez JD. Association between physical activity and changes in intestinal microbiota composition: A systematic review. PLoS One. 2021;16(2):e0247039. doi: 10.1371/journal.pone.0247039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gervasi M, Sisti D, Amatori S, Donati Zeppa S, Annibalini G, Piccoli G, et al. Effects of a commercially available branched-chain amino acid-alanine-carbohydrate-based sports supplement on perceived exertion and performance in high intensity endurance cycling tests. J Int Soc Sports Nutr. 2020;17(1):6. doi: 10.1186/s12970-020-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41(1):e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos P, et al. vegan: Community Ecology Package. 2008.
- 24.Kassambara A. rstatix: Pipe-Friendly Framework for Basic Statistical Tests. 2021.
- 25.Hothorn T, Winell H, Hurnik K, Van de Wiel MA, Zeileis A. coin: Conditional Inference Procedures in a Permutation Test Framework. 2021.
- 26.Tomczak M, Tomczak E. The need to report effect size estimates revisited. An overview of some recommended measures of effect size. Trends in Sport Sciences. 2014;1(21):19–25. [Google Scholar]
- 27.Hamilton N. ggtern: An Extension to 'ggplot2', for the Creation of Ternary Diagrams. 2020.
- 28.Wickham H, Chang W, Henry L, Pedersen TL, Takahashi K, Wilke C, et al. ggplot2: Create Elegant Data Visualisations Using the Grammar of. Graphics. 2020.
- 29.Wei T, Simko V. corrplot: Visualization of a Correlation Matrix. 2021.
- 30.Gu Z. circlize: Circular Visualization. 2021.
- 31.Yuan X, Xu S, Huang H, Liang J, Wu Y, Li C, et al. Influence of excessive exercise on immunity, metabolism, and gut microbial diversity in an overtraining mice model. Scand J Med Sci Sports. 2018;28(5):1541–1551. doi: 10.1111/sms.13060. [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Zhou H, Zhang L, Li R, Luo L, Yu Z, et al. Effects of high-intensity interval training on gut microbiota profiles in 12 months' old ICR mice. J Physiol Biochem. 2020;76(4):539–548. doi: 10.1007/s13105-020-00758-w. [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Liao J, Fang Y, Deng H, Yin H, Shen B, et al. Six-Week Exercise Training With Dietary Restriction Improves Central Hemodynamics Associated With Altered Gut Microbiota in Adolescents With Obesity. Front Endocrinol (Lausanne). 2020;11:569085. doi: 10.3389/fendo.2020.569085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parada Venegas D, De la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong F, Wen X, Yang M, Lai HY, Momma H, Cheng L, et al. Effect of an 8-week Exercise Training on Gut Microbiota in Physically Inactive Older Women. Int J Sports Med. 2020. [DOI] [PubMed]
- 36.Binda C, Lopetuso LR, Rizzatti G, Gibiino G, Cennamo V, Gasbarrini A. Actinobacteria: A relevant minority for the maintenance of gut homeostasis. Dig Liver Dis. 2018;50(5):421–428. doi: 10.1016/j.dld.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Bourriaud C, Robins RJ, Martin L, Kozlowski F, Tenailleau E, Cherbut C, et al. Lactate is mainly fermented to butyrate by human intestinal microfloras but inter-individual variation is evident. J Appl Microbiol. 2005;99(1):201–212. doi: 10.1111/j.1365-2672.2005.02605.x. [DOI] [PubMed] [Google Scholar]
- 38.Karvonen AM, Sordillo JE, Gold DR, Bacharier LB, O'Connor GT, Zeiger RS, et al. Gut microbiota and overweight in 3-year old children. Int J Obes (Lond). 2019;43(4):713–723. doi: 10.1038/s41366-018-0290-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng Q, Li D, He Y, Li Y, Yang Z, Zhao X, et al. Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci Rep. 2019;9(1):13424. doi: 10.1038/s41598-019-49462-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brahe LK, Le Chatelier E, Prifti E, Pons N, Kennedy S, Hansen T, et al. Specific gut microbiota features and metabolic markers in postmenopausal women with obesity. Nutr Diabetes. 2015;5:e159. doi: 10.1038/nutd.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petersen LM, Bautista EJ, Nguyen H, Hanson BM, Chen L, Lek SH, et al. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome. 2017;5(1):98. doi: 10.1186/s40168-017-0320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castellanos N, Diez GG, Antunez-Almagro C, Bressa C, Bailen M, Gonzalez-Soltero R, et al. Key Bacteria in the Gut Microbiota Network for the Transition between Sedentary and Active Lifestyle. Microorganisms. 2020;8(5). [DOI] [PMC free article] [PubMed]
- 43.Hughes RL. A Review of the Role of the Gut Microbiome in Personalized Sports Nutrition. Front Nutr. 2019;6:191. doi: 10.3389/fnut.2019.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frampton J, Murphy KG, Frost G, Chambers ES. Short-chain fatty acids as potential regulators of skeletal muscle metabolism and function. Nat Metab. 2020;2(9):840–848. doi: 10.1038/s42255-020-0188-7. [DOI] [PubMed] [Google Scholar]
- 45.Hawley JA. Microbiota and muscle highway - two way traffic. Nat Rev Endocrinol. 2020;16(2):71–72. doi: 10.1038/s41574-019-0291-6. [DOI] [PubMed] [Google Scholar]
- 46.Nay K, Jollet M, Goustard B, Baati N, Vernus B, Pontones M, et al. Gut bacteria are critical for optimal muscle function: a potential link with glucose homeostasis. Am J Physiol Endocrinol Metab. 2019;317(1):E158–EE71. doi: 10.1152/ajpendo.00521.2018. [DOI] [PubMed] [Google Scholar]
- 47.Przewlocka K, Folwarski M, Kazmierczak-Siedlecka K, Skonieczna-Zydecka K, Kaczor JJ. Gut-Muscle AxisExists and May Affect Skeletal Muscle Adaptation to Training. Nutrients. 2020;12(5). [DOI] [PMC free article] [PubMed]
- 48.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magnusson MK, Isaksson S, Ohman L. The Anti-inflammatory Immune Regulation Induced by Butyrate Is Impaired in Inflamed Intestinal Mucosa from Patients with Ulcerative Colitis. Inflammation. 2020;43(2):507–517. doi: 10.1007/s10753-019-01133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khoshbin K, Camilleri M. Effects of dietary components on intestinal permeability in health and disease. Am J Physiol Gastrointest Liver Physiol. 2020;319(5):G589–G608. doi: 10.1152/ajpgi.00245.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rios-Covian D, Salazar N, Gueimonde M, de Los Reyes-Gavilan CG. Shaping the Metabolism of Intestinal Bacteroides Population through Diet to Improve Human Health. Front Microbiol. 2017;8:376. doi: 10.3389/fmicb.2017.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durk RP, Castillo E, Marquez-Magana L, Grosicki GJ, Bolter ND, Lee CM, et al. Gut Microbiota Composition Is Related to Cardiorespiratory Fitness in Healthy Young Adults. Int J Sport Nutr Exerc Metab. 2019;29(3):249–253. doi: 10.1123/ijsnem.2018-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y, Shi Y, Wiklund P, Tan X, Wu N, Zhang X, et al. The Association between Cardiorespiratory Fitness and Gut Microbiota Composition in Premenopausal Women. Nutrients. 2017;9(8). [DOI] [PMC free article] [PubMed]
- 54.Yu Y, Mao G, Wang J, Zhu L, Lv X, Tong Q, et al. Gut dysbiosis is associated with the reduced exercise capacity of elderly patients with hypertension. Hypertens Res. 2018;41(12):1036–1044. doi: 10.1038/s41440-018-0110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheiman J, Luber JM, Chavkin TA, MacDonald T, Tung A, Pham LD, et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med. 2019;25(7):1104–1109. doi: 10.1038/s41591-019-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mancin L, Rollo I, Mota JF, Piccini F, Carletti M, Susto GA, et al. Optimizing Microbiota Profiles for Athletes. Exerc Sport Sci Rev. 2021;49(1):42–49. doi: 10.1249/JES.0000000000000236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.