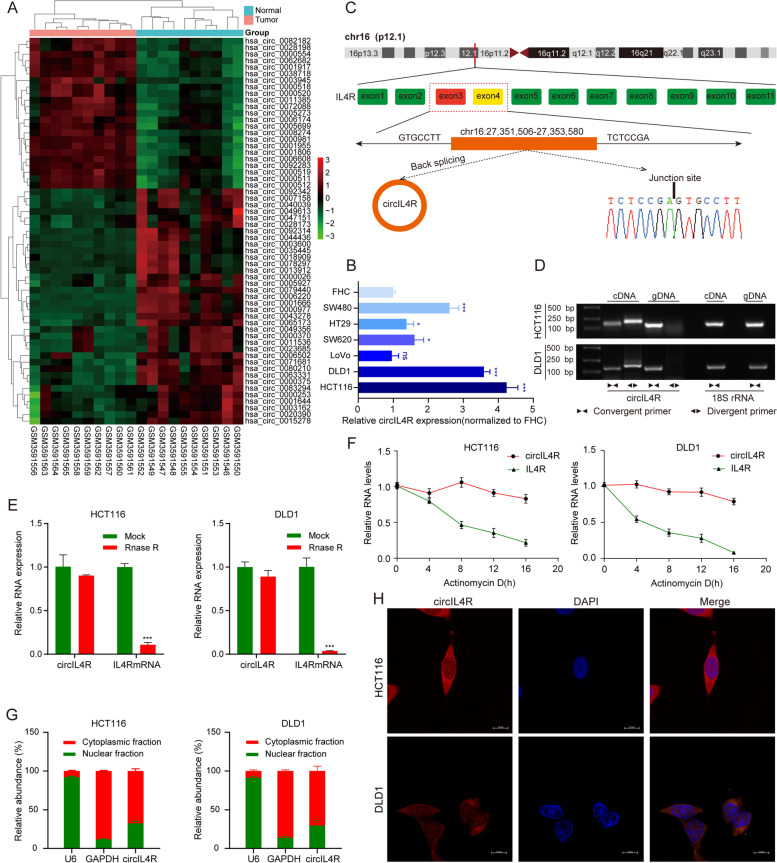
Fig. 1.
Identification and characterization of circIL4R in CRC cells. a Heatmap showing the significantly differentially expressed circRNAs in ten pairs of CRC tissues and corresponding normal tissues (n = 20). Red and green denote upregulated and downregulated circRNAs, respectively. b Relative expression of circIL4R in a normal colorectal epithelium cell line (FHC) and CRC cell lines (HCT116, DLD1, LoVo, SW620, HT29 and SW480). 18S rRNA served as internal reference. c Schematic illustration of the formation of circIL4R via circularization from exons 3 to 4 in the IL4R gene. d The existence of circIL4R was examined by agarose gel electrophoresis with the PCR products. circIL4R could be amplified by divergent primers in the complementary DNA (cDNA) but not genomic DNA (gDNA). 18S rRNA was used as a positive control. e The expression of circIL4R and IL4R mRNA in CRC cells treated with RNase R was examined by qRT-PCR. f The stability of circIL4R and IL4R mRNA in CRC cells treated with actinomycin D at the indicated times was examined by qRT-PCR. g and h qRT-PCR assay of the subcellular fractionation products and representative FISH images showing that the localization of circIL4R in CRC cells was primarily in the cytoplasm. *P < 0.05, **P < 0.01, ***P < 0.001