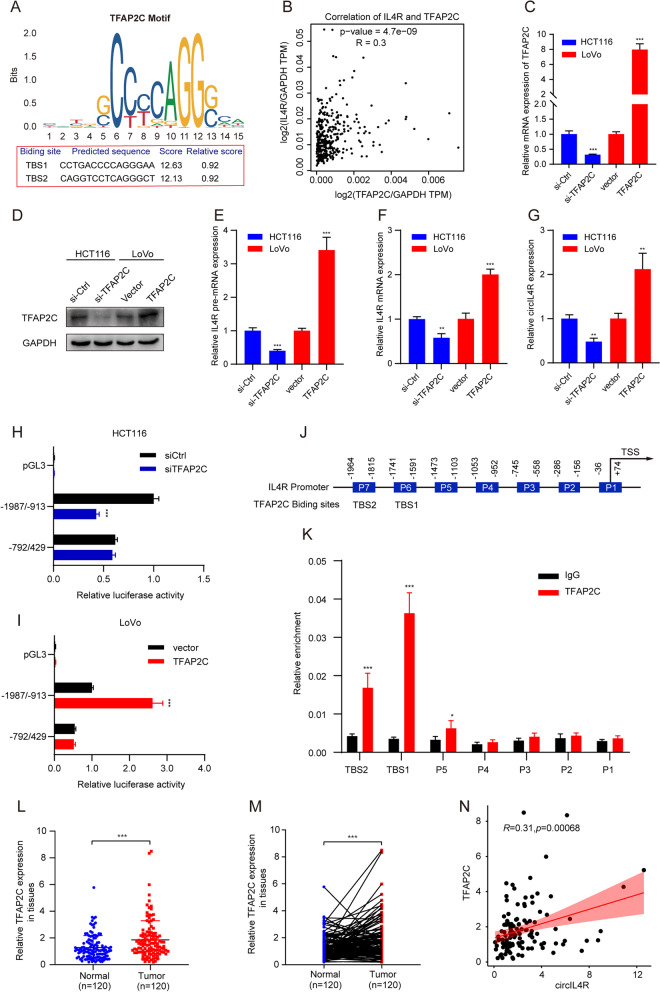
Fig. 3.
TFAP2C enhances circIL4R expression via transcriptional regulation. a The predicted TFAP2C-binding sequence in the IL4R promoter region was identified with the JASPAR website; the scores of the predicted sequences termed TBS1 and TBS2 are shown. b. TFAP2C expression in CRC was positively correlated with IL4R expression based on the analysis of the GEPIA online dataset. c and d qRT-PCR and western blot assays validating the TFAP2C mRNA and protein expression in HCT116 and LoVo cells transfected with siRNAs and overexpression plasmids, respectively. e-g qRT-PCR showed that the expression of pre-mRNA, mRNA and circRNA of IL4R was significantly upregulated in CRC cells transfected with TFAP2C overexpression plasmids but downregulated in CRC cells with TFAP2C knockdown. h and i Luciferase reporter assays of the pGL-IL4R promoter showed that luciferase activity driven by the − 1987/− 913 fragment of the IL4R promoter region was significantly reduced in HCT116 cells with TFAP2C knockdown but enhanced in LoVo cells with TFAP2C overexpression. j Schematic illustration of the seven IL4R promoter regions (named P1–P7) designed for TFAP2C potential binding regions (above). Schematic illustration of predicted TFAP2C binding sites (named TBS1 and TBS2) (below). k The ChIP assay and qRT-PCR analysis showed that TFAP2C bound to the TBS1-TBS2 and P5 regions in the IL4R promoter; IgG was used as the negative control. l and m qRT-PCR analysis showed that TFAP2C mRNA expression was significantly upregulated in 120 CRC tissues compared with that in paired ANTs. n. The mRNA expression of TFAP2C in CRC was positively correlated with circIL4R based on the qRT-PCR results. *P < 0.05, **P < 0.01, ***P < 0.001