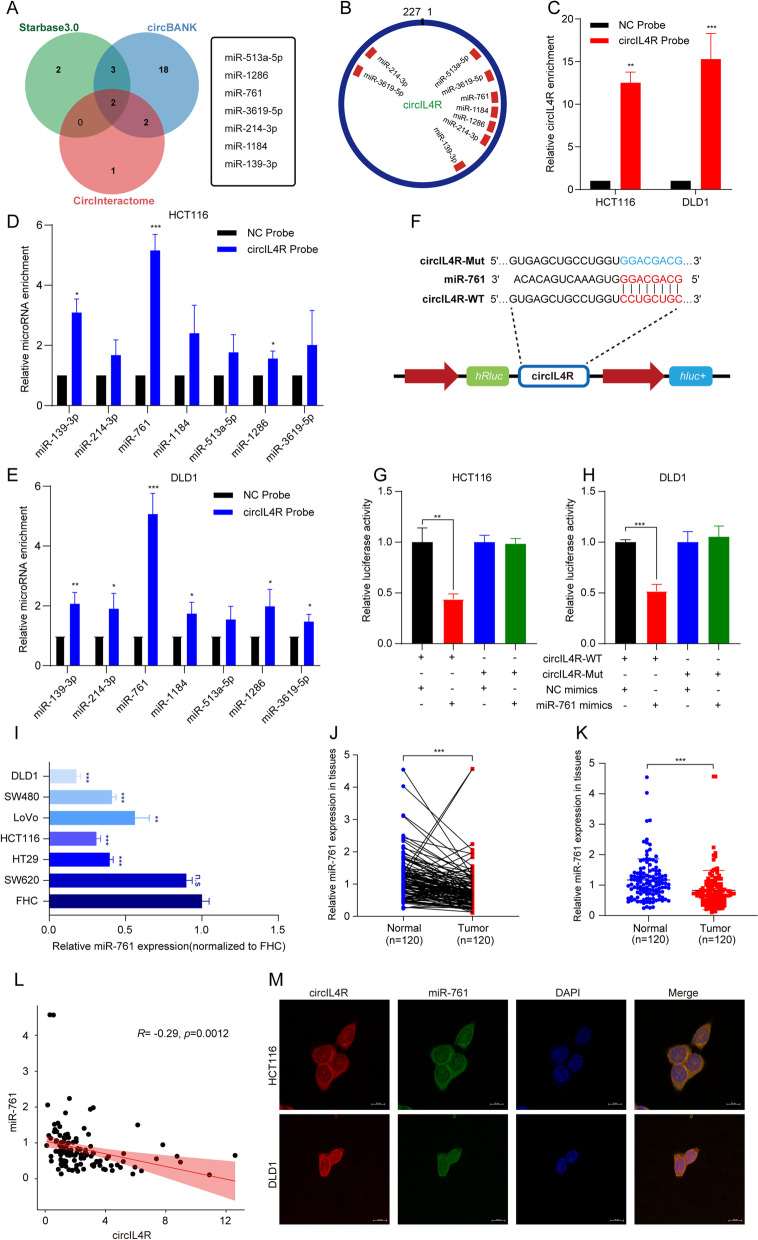
Fig. 6.
circIL4R acts as a sponge for miR-761 in CRC cells. a Seven miRNAs potentially targeted by circIL4R were predicted by cross-analysis with Circular RNA Interactome, circBANK and starbase3.0 databases. b Schematic illustration of the putative binding sites of seven miRNA candidates predicted to bind with circIL4R. c The efficiency of the biotinylated circIL4R probe in CRC cells was validated by qRT-PCR. d and e The relative expression of the miRNA candidates enriched by the circIL4R probe in CRC cells was assessed by qRT-PCR. f Schematic illustration of WT and Mut circIL4R luciferase reporter vectors. g and h CRC cells were cotransfected with miR-761 mimics and either WT or Mut circIL4R luciferase reporter vectors, and the activity of the luciferase reporter was determined. i Relative expression of miR-761 in a normal colorectal epithelium cell line (FHC) and CRC cell lines (HCT116, DLD1, LoVo, SW620, HT29 and SW480). j-l qRT-PCR analysis showed that miR-761 expression was significantly downregulated in 120 CRC tissues compared with paired ANTs and was negatively correlated with circIL4R expression. m Colocalization between circIL4R and miR-761 in CRC cells was verified by FISH staining. DAPI was used to stain the nuclei. Scale bar, 10 μm. *P < 0.05, **P < 0.01, ***P < 0.001