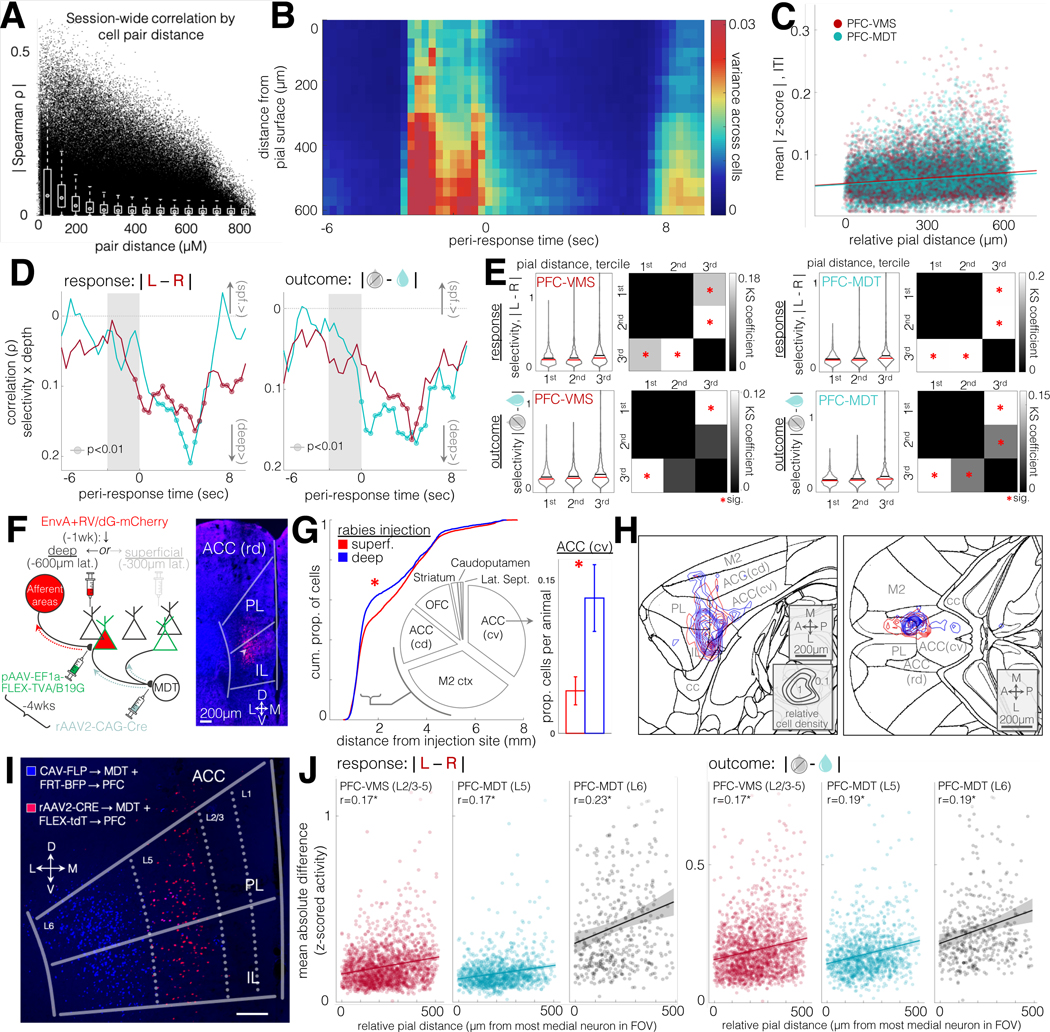
Figure 7: Post-trial feedback-related activity in projection populations is organized by a common topological gradient.
A. Spatial distance of all simultaneously recorded cell pairs vs. temporal correlation. Correlations are partial correlations, controlling for changes in putative neuropil (temporal profile background component from CNMF-E source extraction algorithm). Spearman R for distance vs partial correlation is −0.25, p=0 for N = 1562607 pairs from 4740 cells in 21 animals. White plots are box plots for 60μm bins.
B. Cross-neuron variance of trial-averaged activity, over trial-aligned timepoints and relative distance from pial surface. Warmer colors correspond with higher variance per spatiotemporal bin. Units are means of z-scored activity. Same cells as in (A).
C. Time-averaged data from (B), broken out by neuron and by projection cell type. Correlation of absolute values of z-scored activity with relative pial distance during ITI for PFC-VMS and PFC-MDT neurons. PFC-MDT Spearman ρ=0.16, p=8×10-79, 1155 neurons, 9 animals; PFC-VMS Spearman ρ=0.12, p=3×10-52, N=1770 neurons, 8 animals.
D. Correlations of feature selectivity (mean absolute difference in z-scored activity between conditions) with pial distance at trial-aligned timepoints. Left: response selectivity; right: outcome selectivity. Circles, Spearman p<0.01. PFC-VMS in red, PFC-MDT in green (same units as in D). Note: y-axis is inverted so that “deep>” values appear on the bottom.
E. Distributions of cell feature selectivity for response and outcome during ITI for PFC-VMS and PFC-MDT populations, binned by tercile of relative pial distance. Top row, response selectivity; bottom row, outcome selectivity; left column, PFC-VMS neurons; right column, PFC-MDT neurons. Red cross-bars are medians, black cross-bars are means. Binned spatial depths were tested for significant differences using a Kolmogorov-Smirnov fit test. For trial outcome selectivity across spatial depth, the KS Chi-square for PFC-MDT neurons was 23.09, p=4×10-05; for PFC-VMS neurons, Chi-square was 19.3, p=0.0002. For response selectivity across spatial depth, KS Chi-square for PFC-MDT neurons was 30.2, p=1×10-6; for PFC-VMS neurons, Chi-square 36.54, p=6×10-8. Checkered plots are coefficients from post-hoc 2-sample Kolmogorov-Smirnov tests, comparing distribution similarity across all pairs of terciles. Red asterisks are comparisons with Bonferroni-corrected significance.
F. Left: Schematic diagram of G-deleted rabies tracing experiment (see Methods). Right: example coronal photomicrograph (AP +1.7mm) of Rabies-mCherry-labeled cells at a deep injection site (white arrow). N=45421 cells from 22 animals, 13 deep, 9 superficial injections. Starter cells were PFC-VMS (N=6 injections) and PFC-MDT (N=16 injections). Two-way GLM revealed no significant differences in the number of cells labeled according to projection cell type (t=1, p=0.3) or injection depth (t=1.1, p=0.3).
G. Kolmogorov-Smirnov comparison of cell density distributions by distance from injection site. Traces are cumulative density of cells labeled by deep (blue) and superficial (red) injections (KS p=2.07×10-26, n=9752 and 4755 cells from superficial and deep injections in 9 and 13 animals, respectively). The regions immediately surrounding the injection site (PL, IL, and rostral ACC) were excluded, as they were likely to contain primarily starter cells. The largest difference in labeled cell density between deep and superficial injections was observed in the range of 1mm to 2mm distance from the injection site. We therefore compared counts in all regions containing labeled cells within this range of distances (pie chart inset). Labeled cells in this range were most dense in caudo-ventral ACC (35%), secondary motor cortex (34%), caudo-dorsal ACC (16%), orbitofrontal cortex (combined medial/lateral, 11%), striatum (2%), and caudo-putamen and lateral septum (<1%). Only the caudo-ventral ACC showed a significant difference in cell count by injection depth across animals, with deep injections resulting in a higher proportion of cells in cv-ACC than superficial injections (0.13 ± 0.03 vs 0.04 ± 0.01 of total per animal, t=2.2, p=0.04).
H. Cell density contour plots for sagittal (left) and horizontal (right) cross-sections through the regions surrounding ACC. Concentric contours show summed density pooled across all animals (blue, deep-injection animals, red, superficial-injection animals), normalized within the cross-section shown (a.u.). Higher cv-ACC density in deep-injected neurons is shown in both orientations. Relatively higher density in M2 cortex in superficial-injection animals did not meet significance.
I. Labeled subpopulations of PFC-MDT cells with selective tropism for rAAV2 (predominantly located in layer 5, hereafter referred to as PFC(L5)-MDT neurons) and CAV (predominantly located in layer 6, hereafter PFC(L6)-MDT neurons). Dual labeling was achieved by co-injecting a viral mixture of rAAV2-Cre and CAV2-FLP in MDT, and a second viral mixture of FLEX-tdT and FRT-BFP in PFC.
J. Pial depth and feature selectivity for the three projection subtypes. Left: Kruskal Wallis anova of cell selectivity for response selectivity across cell types: Chi-square statistic = 440, p=2×10-96. Post-hoc GLM testing of cell type by pial distance showed significant group differences between PFC-VMS vs PFC(L6)-MDT (t=3.4, p=0.0007), and for PFC(L5)-MDT vs PFC(L6)-MDT (t=3.9, p=0.0001), but not for PFC-VMS vs PFC(L5)-MDT (t=1.0, p=0.3). Post-hoc testing of within-group effect of pial distance on cell selectivity for response and trial outcome revealed parallel effects (no cell-type-by-pial-distance interaction effects) for all three groups. Spearman R for pial distance by response selectivity was significant for PFC-VMS (r=0.17, p=1.9×10-13), PFC(L5)-MDT (r=0.17, p=6.6×10-9), and PFC(L6)-MDT (r=0.23, p=1.8×10-6). Right: Kruskal Wallis non-parametric anova of cell selectivity for trial outcome (mean absolute difference in z-scored activity between correct and incorrect trials during the subsequent inter-trial interval) across cell types: Chi-square statistic = 134, p=1×10-30. Post-hoc GLM testing of cell type by pial distance, however, showed no group differences between PFC-VMS vs PFC(L5)-MDT (t=0.5, p=0.6), between PFC-VMS and PFC(L6)-MDT (t=1.25, p=0.2), or between PFC(L5)-MDT and PFC(L6)-MDT (t=1.6, p=0.1). Spearman R for pial distance by trial outcome selectivity was also significant for each group: PFC-VMS (r=0.17, p=1.4×10-13), PFC(L5)-MDT (r=0.19, p=8.1×10-11), and PFC(L6)-MDT (r=0.19, p=8.3×10-5). PFC-VMS: N=1155 cells from 8 animals; PFC(L5)-MDT: N=1770 cells from 9 animals; PFC(L6)-MDT: N=430 cells from 8 animals.