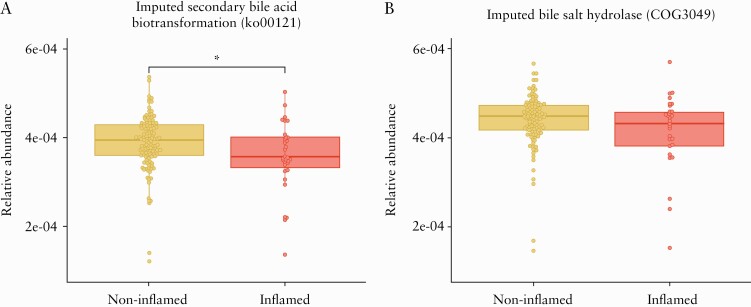
Figure 1.
Inferred bile acid-metabolising gene abundance in terminal ileum samples [n = 140 samples]. Data expressed as relative abundance of imputed gene function from 16S marker gene data using PICRUSt. Differential abundance analysis was performed by linear regression modelling including endoscopic inflammation [non-inflamed and inflamed], IBD type [CD and UC/IBDU], sex, and age as covariates. A] Reduced relative abundance of ko00121 pathway gene abundance is associated with endoscopic inflammation. B] A non-significant numerical reduction of cluster of orthologous groups [COG3049] in inflamed samples was observed; *p-value <0.05 for the linear regression analysis. UC, ulcerative colitis; IBDU, inflammatory bowel disease unclassified; CD, Crohn´s disease.