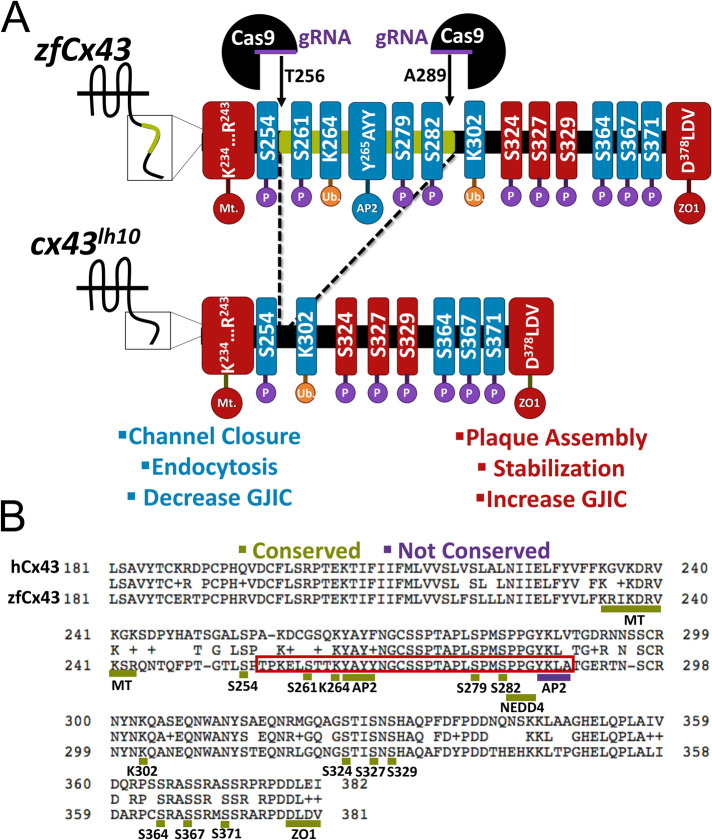
FIGURE 1:
Generation of the cx43lh10 zebrafish mutant using the CRISPR/Cas9 gene-editing tool. (A) Schematic of the zebrafish WT Cx43 CT (top) and cx43lh10 CT (bottom). Two gRNAs were designed, one that targeted amino acid 256 and the other that targeted amino acid 289. After Cas9-mediated generation of double strand breaks, cells employ nonhomologous end joining to repair the breaks, omitting amino acids 256–289 from the Cx43 gene. The portion of Cx43 deleted in cx43lh10 (green) contains most of the key residues involved in GJ endocytosis including the conserved AP2/clathrin binding site, critical MAPK phosphorylation sites on serine residues 261 and 279/282, and one of the two polyubiquitination sites (blue), whereas residues known to be involved in plaque assembly, stabilization, and increased GJIC (red) are left intact. (B) Amino acid sequence alignment of the CT of human Cx43 (top) and zebrafish Cx43 (bottom). The region boxed in red encompasses the deleted amino acid residues and displays the critical endocytosis-related amino acid motifs. Note that except for the second AP-2/clathrin binding site (S3, underlined blue), endocytic motifs identified in mammalian Cx43 are conserved in zebrafish Cx43 (underlined green). All residues in A and B are labeled following zebrafish nomenclature, which is partially shifted one position to the left compared with mammalian Cx43 due to the deletion of one amino acid residue in the CT of zebrafish Cx43 at position 251.