Abstract
Leaf senescence is the last stage of leaf development and is an orderly biological process accompanied by degradation of macromolecules and nutrient recycling, which contributes to plant fitness. Forward genetic mutant screening and reverse genetic studies of senescence-associated genes (SAGs) have revealed that leaf senescence is a genetically regulated process, and the initiation and progression of leaf senescence are influenced by an array of internal and external factors. Recently, multi-omics techniques have revealed that leaf senescence is subjected to multiple layers of regulation, including chromatin, transcriptional and post-transcriptional, as well as translational and post-translational levels. Although impressive progress has been made in plant senescence research, especially the identification and functional analysis of a large number of SAGs in crop plants, we still have not unraveled the mystery of plant senescence, and there are some urgent scientific questions in this field, such as when plant senescence is initiated and how senescence signals are transmitted. This paper reviews recent advances in the multiple layers of regulation on leaf senescence, especially in post-transcriptional regulation such as alternative splicing.
Keywords: leaf senescence, senescence-associated genes, multi-omics, gene regulatory network, alternative splicing
Introduction
Plant leaves are the main organ for photosynthesis, converting light energy into chemical energy stored in carbohydrate molecules, which is the main source of energy for all organisms on earth. Senescence is the final stage of leaf development process, which is a slow and complex biological process including the initiation, progression, and terminal phases (Guo and Gan, 2005; Lim et al., 2007). The degradation of chlorophyll and chloroplasts occurs in the later phase of leaf senescence, accompanied by the degradation of macromolecules such as proteins, lipids, and nucleic acids. In annual plants, the nutrients released from senescent leaves are transferred to actively growing young leaves and seeds to increased reproductive success. In perennial plants such as deciduous trees, the nitrogen from leaf proteins is relocated to form bark storage proteins (BSP) in phloem tissues, and then remobilized and reutilized for spring shoot growth. Therefore, the timing of leaf senescence plays an important role in ensuring nutrient recycling, adaptation to the environment, and reproduction in plants. A number of studies in crops such as wheat and rice revealed that alteration of leaf senescence process could significantly affect the yield and quality of crops. Extended lifespan of leaves in apple trees greatly improved fruit quality in apple trees (Han et al., 2020; Hu et al., 2020), and increased fruit yield and sugar content in tomato (Solanum lycopersicon) (Lira et al., 2017; Ma et al., 2018). Moreover, delayed leaf senescence conferred enhance drought resistance in tobacco or cassava (Zhang et al., 2010). Therefore, an in-depth understanding of the regulatory mechanisms of leaf senescence is of great importance.
Leaf senescence is not a passive and disorderly process, but a highly programmed degenerative process (Guo and Gan, 2005). The initiation and progression of leaf senescence are influenced by numerous endogenous developmental signals and external environmental factors. Leaf age is the most important endogenous cue that determines the initiation of leaf senescence. However, the nature of age and how age information is perceived remains a mystery (Jing et al., 2002). Plant hormones such as ethylene, jasmonic acid (JA), salicylic acid (SA), abscisic acid (ABA), brassinosteroid (BR), and strigolactone (SL) promote leaf senescence and are extensively involved in response to various abiotic and biotic stresses, whereas auxin, cytokinins (CKs), and gibberellins (GAs) delay leaf senescence (Lim et al., 2007; Miao and Zentgraf, 2007; Li et al., 2013; Zhang et al., 2013; Yamada and Umehara, 2015; Kim et al., 2020). Hormone signaling pathways often mediate or influence development and environmental responses to regulate leaf senescence (Lim et al., 2007). Interestingly, changes in the circadian rhythm of plants also impact leaf senescence, but the causal relationship between them needs to be further explored (Song et al., 2018). In addition to being regulated by plant age or phytohormones, leaf senescence can also be caused by numerous environmental stresses such as darkness, nutrient deficiency, drought stress, and pathogen infection (Lim et al., 2007; Chen et al., 2013; Woo et al., 2019; Li et al., 2020b). There is much information about age- or abiotic stress-induced leaf senescence, whereas little is known about the molecular basis of biotic stress-triggered senescence. Recently, it was found that the secretory effector protein PevD1 (Protein elicitor from Verticillium. dahliae 1) plays an important role in the V. dahliae–induced senescence process. PevD1 interacts with ORESARA1 (ORE1), one core transcription factor regulating plant senescence (Lim et al., 2007), and attenuates the NLA-mediated degradation of ORE1, thereby enhancing ethylene biosynthesis by directly binding the promoter of 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID (ACC) SYNTHASE 6 (ACS6) (Zhang Y. et al., 2021). This research provides a mechanism for previous observations that ethylene contributes to V. dahliae–induced premature leaf senescence.
Multiple-Layers of Regulation on Leaf Senescence
In the past few decades, remarkable progress has been made in leaf senescence research, and time-evolving genetic networks have been established through genetics and multi-omics strategies, allowing us to gain a deeper understanding of this important biological process (Kim H. J. et al., 2018).
Here, we reviewed the recent advances in the molecular regulation of leaf senescence, including chromatin level, transcription level, as well as post-transcriptional, translational, and post-translational level (Figure 1). We also summarized the key players involved in the multilevel regulation of leaf senescence (Table 1).
FIGURE 1.
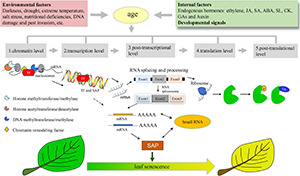
Multiple Layers of Regulation on Plant Leaf Senescence. Plant leaf senescence is finely regulated by endogenous developmental signals, and external environmental cues in an age-dependent manner. The multiple layers of regulation on leaf senescence includes epigenetic regulation at the chromatin level involved in histone proteases, DNA methylation modifying enzymes and chromatin remodeling factors, the master transcription factors involved in transcription regulation such as WRKYs and NACs, also includes miRNA and alternative splicing involved in the post-transcriptional level, and translation initiation and elongation factors involved in the regulation of translation level, as well as post-translational ubiquitination and phosphorylation. SAP, senescence-associated protein; Ub, ubiquitin; TF, transcription factors.
TABLE 1.
List of the key genes involved in multiple layers of regulation on leaf senescence.
| Gene | Species | Effects | Regulation | References |
| AtHD1 | Arabidopsis thaliana | Delay | Chromatin level | Pandey et al., 2002 |
| AtSRT1 | Arabidopsis thaliana | Delay | Chromatin level | Liu et al., 2017 |
| BRAHMA | Arabidopsis thaliana | Delay | Chromatin level | Efroni et al., 2013 |
| DRD1/DDM1 | Arabidopsis thaliana | Promote | Chromatin level | Cho et al., 2016 |
| HAC1 | Arabidopsis thaliana | Promote | Chromatin level | Hinckley et al., 2019 |
| HDA9 | Arabidopsis thaliana | Delay | Chromatin level | Zheng et al., 2016 |
| HDA15 | Arabidopsis thaliana | Delay | Chromatin level | Shen et al., 2019 |
| HDA19 | Arabidopsis thaliana | Delay | Chromatin level | Ueda et al., 2018 |
| HD2C | Arabidopsis thaliana | Delay | Chromatin level | Buszewicz et al., 2016 |
| JMJ16 | Arabidopsis thaliana | Promote | Chromatin level | Liu et al., 2019 |
| SUVH2 | Arabidopsis thaliana | Delay | Chromatin level | Ay et al., 2009 |
| REF6 | Arabidopsis thaliana | Promote | Chromatin level | Wang et al., 2019 |
| WRKY6 | Arabidopsis thaliana | Promote | Transcription level | Robatzek and Somssich, 2001 |
| WRKY22 | Arabidopsis thaliana | Promote | Transcription level | Zhou et al., 2011 |
| WRKY42 | Arabidopsis thaliana | Promote | Transcription level | Niu et al., 2020 |
| WRKY45 | Arabidopsis thaliana | Promote | Transcription level | Chen et al., 2017 |
| WRKY46 | Arabidopsis thaliana | Promote | Transcription level | Zhang D. et al., 2021 |
| WRKY53 | Arabidopsis thaliana | Promote | Transcription level | Zentgraf et al., 2010 |
| WRKY54/WRKY70 | Arabidopsis thaliana | Delay | Transcription level | Besseau et al., 2012 |
| WRKY55 | Arabidopsis thaliana | Promote | Transcription level | Wang et al., 2020 |
| WRKY57 | Arabidopsis thaliana | Delay | Transcription level | Jiang et al., 2014 |
| WRKY75 | Arabidopsis thaliana | Promote | Transcription level | Guo P. et al., 2017 |
| GhWRKY42 | Gossypium hirsutum | Promote | Transcription level | Gu et al., 2018 |
| GhWRKY91 | Gossypium hirsutum | Delay | Transcription level | Gu et al., 2019 |
| CpWRKY71 | Chimonanthus praecox | Promote | Transcription level | Huang et al., 2019 |
| OsWRKY42 | Oryza sativa | Promote | Transcription level | Han et al., 2014 |
| OsWRKY93 | Oryza sativa | Promote | Transcription level | Li Y. et al., 2021 |
| BnaWSR1 | Brassica napus | Promote | Transcription level | Cui et al., 2020 |
| BnaWGR1 | Brassica napus | Promote | Transcription level | Yang et al., 2018 |
| BrWRKY6 | Brassica rapa var. parachinensis | Promote | Transcription level | Fan et al., 2018 |
| AtNAP | Arabidopsis thaliana | Promote | Transcription level | Guo and Gan, 2006 |
| AtNAC3 | Arabidopsis thaliana | Promote | Transcription level | Hickman et al., 2013 |
| ATAF1 | Arabidopsis thaliana | Promote | Transcription level | Garapati et al., 2015 |
| ANAC016 | Arabidopsis thaliana | Promote | Transcription level | Kim et al., 2013 |
| ANAC017, VNI1, and ANAC090 | Arabidopsis thaliana | Delay | Transcription level | Woo et al., 2016 |
| ANAC019 | Arabidopsis thaliana | Promote | Transcription level | Lee et al., 2015 |
| ANAC032 | Arabidopsis thaliana | Promote | Transcription level | Mahmood et al., 2016 |
| ANAC046 | Arabidopsis thaliana | Promote | Transcription level | Oda-Yamamizo et al., 2016 |
| ANAC072 | Arabidopsis thaliana | Promote | Transcription level | Li S. et al., 2016 |
| NAC075 | Arabidopsis thaliana | Delay | Transcription level | Kan et al., 2021 |
| ANAC102 | Arabidopsis thaliana | Promote | Transcription level | Nakashima et al., 2012 |
| JUB1 | Arabidopsis thaliana | Delay | Transcription level | Wu et al., 2012 |
| NTL9 | Arabidopsis thaliana | Promote | Transcription level | Yoon et al., 2008 |
| ORE1 | Arabidopsis thaliana | Promote | Transcription level | Kim et al., 2009 |
| ORS1 | Arabidopsis thaliana | Promote | Transcription level | Balazadeh et al., 2011 |
| PIF4 and PIF5 | Arabidopsis thaliana | Promote | Transcription level | Sakuraba et al., 2014; Song et al., 2014 |
| VNI2 | Arabidopsis thaliana | Delay | Transcription level | Yang et al., 2011 |
| BnaNAC87 | Brassica napus | Promote | Transcription level | Yan et al., 2018 |
| lbNAC1 | Ipomoea batatas | Promote | Transcription level | Chen et al., 2016 |
| MpSNAC67 | Musa x paradisiaca | Promote | Transcription level | Tak et al., 2018 |
| MlNAC5 | Miscanthus lutarioriparius | Promote | Transcription level | Yang et al., 2015 |
| NtNAC080 | Nicotiana tabacum | Promote | Transcription level | Li et al., 2018 |
| NAM-B1 | Triticum turgidum | Promote | Transcription level | Uauy et al., 2006 |
| TaNAC-S | Triticum turgidum | Delay | Transcription level | Zhao et al., 2015 |
| ONAC011 | Oryza sativa | Promote | Transcription level | El Mannai et al., 2017 |
| ONAC096 | Oryza sativa | Delay | Transcription level | Kang et al., 2019 |
| ONAC106 | Oryza sativa | Delay | Transcription level | Sakuraba et al., 2015 |
| OsNAC2 | Oryza sativa | Promote | Transcription level | Mao et al., 2017 |
| OsNAP | Oryza sativa | Promote | Transcription level | Liang et al., 2014 |
| OsDOS | Oryza sativa | Delay | Transcription level | Kong et al., 2006 |
| SlNAP1 and SlNAP2, | Solanum lycopersicon | Promote | Transcription level | Ma et al., 2018 |
| SlORE1S06, SlORE1S03, and SlORE1S02 | Solanum lycopersicon | Promote | Transcription level | Lira et al., 2017 |
| SiNAC1 | Setaria italica | Promote | Transcription level | Ren et al., 2018 |
| GmNAC065 and GmNAC085 | Glycine max | Promote | Transcription level | Melo et al., 2018 |
| GmNAC81 | Glycine max | Promote | Transcription level | Pimenta et al., 2016 |
| GhNAC12 | Gossypium hirsutum | Promote | Transcription level | Zhao et al., 2016 |
| AIF2 | Arabidopsis thaliana | Delay | Transcription level | Kim et al., 2020 |
| AP2 | Arabidopsis thaliana | Delay | Transcription level | Balanza et al., 2018 |
| bHLH03, bHLH13, bHLH14, and bHLH17 | Arabidopsis thaliana | Delay | Transcription level | Qi et al., 2015 |
| CRF1, CRF2, CRF3, CRF5, and CRF6 | Arabidopsis thaliana | Promote | Transcription level | Raines et al., 2016 |
| CBF2 | Arabidopsis thaliana | Delay | Transcription level | Sharabi-Schwager et al., 2010 |
| DEAR1 | Arabidopsis thaliana | Promote | Transcription level | Tsutsui et al., 2009 |
| FYF | Arabidopsis thaliana | Delay | Transcription level | Chen et al., 2011 |
| IAA17 | Arabidopsis thaliana | Promote | Transcription level | Shi et al., 2015 |
| KHZ1 and KHZ2 | Arabidopsis thaliana | Promote | Transcription level | Yan et al., 2017 |
| MYC2, MYC3, and MYC4 | Arabidopsis thaliana | Promote | Transcription level | Qi et al., 2015 |
| MYC5 | Arabidopsis thaliana | Promote | Transcription level | Song et al., 2017 |
| JAZ7 | Arabidopsis thaliana | Promote | Transcription level | Yu et al., 2016 |
| MdbHLH3 | Malusdomestica | Promote | Transcription level | Hu et al., 2020 |
| AtMYBL | Arabidopsis thaliana | Promote | Transcription level | Zhang et al., 2011 |
| MYB2 | Arabidopsis thaliana | Promote | Transcription level | Buchanan-Wollaston et al., 2005 |
| MYBR1 | Arabidopsis thaliana | Delay | Transcription level | Jaradat et al., 2013 |
| MYBH | Arabidopsis thaliana | Promote | Transcription level | Huang et al., 2015 |
| ORE15 | Arabidopsis thaliana | Delay | Transcription level | Kim J. H. et al., 2018 |
| OsMYC2 | Oryza sativa | Promote | Transcription level | Uji et al., 2017 |
| OsMYB102 | Oryza sativa | Delay | Transcription level | Piao et al., 2019 |
| Rap2.4f | Arabidopsis thaliana | Promote | Transcription level | Xu et al., 2010 |
| REVOLUTA | Arabidopsis thaliana | Promote | Transcription level | Xie Y. et al., 2014 |
| RAV1 | Arabidopsis thaliana | Promote | Transcription level | Woo et al., 2010 |
| ScMYB2S1 | Sugarcane | Promote | Transcription level | Guo X. et al., 2017 |
| SlERF36 | Solanum lycopersicon | Promote | Transcription level | Upadhyay et al., 2013 |
| SUB1A | Oryza sativa | Delay | Transcription level | Fukao et al., 2012 |
| SlFYFL | Solanum lycopersicon | Delay | Transcription level | Xie Q. et al., 2014 |
| SlMBP11 | Solanum lycopersicon | Delay | Transcription level | Guo X. et al., 2017 |
| TCP2, TCP4, and TOP10 | Arabidopsis thaliana | Promote | Transcription level | Schommer et al., 2008 |
| CFM4 | Arabidopsis thaliana | Promote | Post-transcriptional level | Lee et al., 2014 |
| ERF4 | Arabidopsis thaliana | Promote | Post-transcriptional level | Koyama et al., 2013; Riester et al., 2019 |
| miR156 | Arabidopsis thaliana | Delay | Post-transcriptional level | Wang, 2014 |
| miR164 | Arabidopsis thaliana | Delay | Post-transcriptional level | Kim et al., 2009 |
| miR172 | Zea mays | Delay | Post-transcriptional level | Wu et al., 2016 |
| miR319 | Arabidopsis thaliana | Promote | Post-transcriptional level | Schommer et al., 2008 |
| miR840 | Arabidopsis thaliana | Delay | Post-transcriptional level | Yujun et al., 2019 |
| ONAC054 | Oryza sativa | Promote | Post-transcriptional level | Sakuraba et al., 2020 |
| PtRD26 | Populus | Delay | Post-transcriptional level | Wang et al., 2021 |
| SlymiR208 | Solanum lycopersicon | Promote | Post-transcriptional level | Zhang Y. et al., 2020 |
| ScMYB2 | Saccharum officinarum | Promote | Post-transcriptional level | Guo X. et al., 2017 |
| u11-48k | Arabidopsis thaliana | Promote | Post-transcriptional level | Xu et al., 2016 |
| eIF5A | Picrorhiza kurrooa | Delay | Translation level | Parkash et al., 2014 |
| LreEF1A4 | Petunia hybrida | Delay | Translation level | Sun et al., 2020 |
| SPL33 | Oryza sativa | Delay | Translation level | Wang et al., 2017 |
| AtSARK | Arabidopsis thaliana | Promote | Post-translational level | Xu et al., 2011 |
| AtWAKL10 | Arabidopsis thaliana | Delay | Post-translational level | Li L. et al., 2021 |
| ATG4a/4b | Arabidopsis thaliana | Delay | Post-translational level | Yoshimoto et al., 2004 |
| ATG9 | Arabidopsis thaliana | Delay | Post-translational level | Hanaoka et al., 2002 |
| ATG10 | Arabidopsis thaliana | Delay | Post-translational level | Phillips et al., 2008 |
| ATG18a | Arabidopsis thaliana | Delay | Post-translational level | Xiong et al., 2005 |
| EDR1 | Arabidopsis thaliana | Delay | Post-translational level | Frye et al., 2001 |
| MKK4/5,MPK1/2 | Arabidopsis thaliana | Promote | Post-translational level | Zhang Y. et al., 2020 |
| MAPKKK18 | Arabidopsis thaliana | Promote | Post-translational level | Matsuoka et al., 2015 |
| MPK6/MKK9 | Arabidopsis thaliana | Promote | Post-translational level | Zhou et al., 2009 |
| GmSARK | Glycine max | Promote | Post-translational level | Li et al., 2006 |
| OsMAPKKK1 | Oryza sativa | Promote | Post-translational level | Wang et al., 2015 |
| PUB12/PUB13 | Arabidopsis thaliana | Delay | Post-translational level | Zhou et al., 2015 |
| RPN10 | Arabidopsis thaliana | Promote | Post-translational level | Lin et al., 2011 |
| RPN5a | Arabidopsis thaliana | Promote | Post-translational level | Book et al., 2009 |
| SAUR49 | Arabidopsis thaliana | Promote | Post-translational level | Wen et al., 2020 |
| SERK4 | Arabidopsis thaliana | Delay | Post-translational level | Li et al., 2019 |
| UPL5 | Arabidopsis thaliana | Delay | Post-translational level | Zentgraf et al., 2010 |
| UBP12/UBP13 | Arabidopsis thaliana | Promote | Post-translational level | Park et al., 2019 |
| UBA2 | Arabidopsis thaliana | Promote | Post-transcriptional level | Kim et al., 2008 |
Chromatin Level
In eukaryotic cells, DNA is packaged into chromatin and its functional units are nucleosomes. The basic unit of chromatin is the nucleosome core particle, a structure in which ∼146 bp of DNA is wrapped around a protein octamer consisting of two subunits each of core histones H2A, H2B, H3, and H4 (Luger et al., 1997; Davey et al., 2002; Marino-Ramirez et al., 2005). The globular region of the histone forms the core of the nucleosome, while the N-terminal tail protrudes from the nucleosomes and is enriched for various post-translational modifications (PTMs), including acetylation, methylation, phosphorylation, and ubiquitination (Bannister and Kouzarides, 2011). These modifications have important regulatory roles, including gene repression, gene activation, and replication (Kouzarides, 2007; Morgan and Shilatifard, 2020). Histone modifications and the enzymes that implement them can facilitate chromatin compaction, nucleosome dynamics, and transcription. These modifications can respond to intrinsic and external stimuli (Kouzarides, 2007). Dysregulation of these processes can alter the balance of gene expression and thus is often observed in many human diseases or plant development, either by gain or loss of function, overexpression, repression through promoter hypermethylation, chromosomal translocation, or mutation of histone-modifying enzymes/complexes, or even histone modification sites (Zhao and Shilatifard, 2019).
Previous investigations revealed that epigenetic modification participates in the plant leaf senescence process. Chromatin immunoprecipitation sequencing (ChIP-seq) analysis using the trimethylation of histone H3 at lysine 4 (H3K4me3) and the trimethylation of histone H3 at lysine 27 (H3K27me3) antibodies reveals the relationship between histone modifications and leaf senescence in Arabidopsis (Brusslan et al., 2012, 2015). Mutation of histone deacetylase AtHD1, a histone modification-related gene, altered leaf senescence process in Arabidopsis (Pandey et al., 2002). The histone acetylation status of specific parts of chromatin is determined by histone acetylases (HATs) and histone deacetylases (HDACs) and their relative activities. Histone acetyltransferase 1 (HAC1) promotes leaf senescence by targeting Ethylene-responsive transcription factor ERF022, a positive regulator of leaf senescence (Hinckley et al., 2019). HISTONE DEACETYLASE 9 (HDA9), HDA15, HDA19, HISTONE DEACETYLASE 2C (HD2C), and SIRTUIN 1 (SRT1) play a potential role in promoting leaf senescence (Buszewicz et al., 2016; Zheng et al., 2016; Liu et al., 2017; Ueda et al., 2018; Hu et al., 2019; Shen et al., 2019). ChIP-seq and fluorescence in situ hybridization (FISH) reveal that the chromatin structure changes as the leaf ages in Arabidopsis. Overexpression of SUVH2, a SU(VAR)3-9 (KMTase1) histone methyltransferase gene, delayed leaf senescence by increasing the dimethylation of histone H3 at lysine 27 (H3K27me2) and H3K27me3 levels in the promoter region of WRKY53, a master positive regulator of leaf senescence, and then suppressing its transcripts (Ay et al., 2009). JUMONJI DOMAIN-CONTAINING PROTEIN 16 (JMJ16), a specific H3K4 demethylase containing JmjC-domain, regulates negatively leaf senescence through repressing the expression of WRKY53 and SENESCENCE-ASSOCIATED GENE 201 (SAG201), two positive regulators of leaf senescence in Arabidopsis. Moreover, genome-wide analysis reveals widespread hypermethylation of H3K4me3 at JMJ16 binding genes, including WRKY53 and SAG201, and coordinated upregulation of their expression in the jmj16 mutant compared with the wild type (Liu et al., 2019). To screen the upstream regulator of NONYELLOWING1 (NYE1) that regulates chlorophyll degradation during leaf senescence by Yeast one-Hybrid (Y1H) approach, the histone H3K27me3 demethylase RELATIVE OF EARLY FLOWERING6 (REF6) was found to directly interact with the NYE1/2 promoter through its zinc finger domain and up-regulates gene expression of positive regulators of leaf senescence such as ETHYLENE INSENSITIVE 2 (EIN2) and ORE1 (Wang et al., 2019). In addition, overexpression of SUVH2 also inhibits the gene expressions of senescence-associated WRKY (Sen-WRKY) and Sen-NAC (NAM/ATAF/CUC) transcription factors, central components of the leaf senescence process, in Arabidopsis leaves upon treatment with bleomycin (BLM), a genotoxic chemical that induces double-strand breaks (DSBs) (Li et al., 2020a).
ATP-dependent chromatin remodeling enzyme involved in chromatin remodeling is also associated with leaf senescence. Mutations of DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1) and DECREASED DNA METHYLATION 1 (DDM1), two SWI2/SNF2 chromatin remodeling proteins, delay leaf senescence (Cho et al., 2016). In contrast, loss-of-function of BRAHMA (BRM), another SWI/SNF2 chromatin remodeling ATPase (Archacki et al., 2017), accelerates leaf senescence (Efroni et al., 2013; Li C. et al., 2016). Further studies are needed in the future to reveal how various epigenetic modifications coordinately regulate leaf senescence.
Transcription Level
Large-scale reprogramming of the transcriptome is a core step in plant leaf senescence. Approximately a dozen percent of genes are up-regulated or down-regulated during leaf senescence in Arabidopsis. Furthermore, master TFs-mediated transcriptional regulation plays a crucial role in the regulation of leaf senescence (Zentgraf et al., 2010; Breeze et al., 2011). A WRKY transcription factor is one of the plant-specific TF families controlling the leaf senescence process. Members of WRKY TFs, including WRKY6, WRKY22, WRKY42, WRKY45, WRKY46, WRKY53, WRKY54, WRKY55, WRKY57, WRKY70, and WRKY75, coordinate with endogenous hormones to finely regulate the leaf senescence process (Robatzek and Somssich, 2001; Miao et al., 2004; Zentgraf et al., 2010; Zhou et al., 2011; Besseau et al., 2012; Jiang et al., 2014; Chen et al., 2017; Guo P. et al., 2017; Niu et al., 2020; Wang et al., 2020). Recently, WRKY46 was found to interact with Non-expressor of Pathogenesis-Related gene 1 (NPR1) and combined with the WRKY6 promoter to induce its expression in response to SA signals, thereby establishing an NPR1-WRKY46-WRKY6 signaling cascade to regulate leaf senescence (Zhang D. et al., 2021). Several WRKY TFs coordinate leaf growth and senescence in plants, including GhWRKY42 and GhWRKY91 in Gossypium hirsutum, BrWRKY6 in cabbage (Brassica rapa), CpWRKY71 in Chimonanthus praecox, OsWRKY93 in Oryza sativa, BnaWSR1 (WRKY regulating SA and ROS 1) and BnaWGR1 (WRKY generating ROS 1) in Brassica napus (Fan et al., 2018; Gu et al., 2018, 2019; Yang et al., 2018; Huang et al., 2019; Cui et al., 2020; Li Y. et al., 2021).
NAC family is one of the largest gene families in plants and plays a central role in regulating leaf senescence. NAC TFs function as positive regulators of leaf senescence, including ORE1/ANAC092, AtNAP/ANAC029, ORE1 SISTER1 (ORS1/ANAC059), ATAF2/ANAC081, ATAF1/ANAC002, ANAC019, AtNAC3/ANAC055, RESPONSIVE TO DESICCATION 26 (ATRD26/ANAC072), ANAC102, ANAC032, ANAC046, ANAC016, and NAC TRANSCRIPTION FACTOR-LIKE 9 (NTL9) or negative regulators such as JUNGBRUNNEN1 (JUB1/ANAC042), ANAC017, VND-INTERACTING1 (VNI1/ANAC082), VND-INTERACTING2 (VNI2/ANAC083), ANAC090, and ANAC075 (Guo and Gan, 2006; Yoon et al., 2008; Kim et al., 2009, 2013; Balazadeh et al., 2011; Nakashima et al., 2012; Wu et al., 2012; Hickman et al., 2013; Garapati et al., 2015; Lee et al., 2015; Takasaki et al., 2015; Kim H. J. et al., 2016, 2018; Li S. et al., 2016; Mahmood et al., 2016; Oda-Yamamizo et al., 2016; Woo et al., 2016; Nagahage et al., 2020; Kan et al., 2021). Future studies need to investigate whether there is communication between these positive and negative regulatory NAC-TFs, which will help to gain insight into the fine regulatory mechanisms of leaf senescence. On the one hand, to investigate whether there are direct interactions between these factors and whether they are synergistic or antagonistic to each other. On the other hand, ChIP-Seq data should be used to analyze whether their target genes overlap to develop a gene regulatory network of leaf senescence. Indeed, some studies have already started to address this aspect. For example, members of NAC-TFs and WRKY-TFs have been found to interact with each other to change the expression of downstream target genes, which in turn triggers leaf senescence (Kim et al., 2009; Zentgraf et al., 2010; Balazadeh et al., 2011; Besseau et al., 2012; Kim H. J. et al., 2016, 2018). Since the process of leaf senescence is accompanied by nutrient return, genes that regulate senescence are likely to regulate crop yield. In supporting this hypothesis, several NAC-TFs regulate crop yield by fine-tuning the initiation and progression of leaf senescence, such as NAM-B1 and TaNAC-S in wheat (Uauy et al., 2006; Zhao et al., 2015; Kang et al., 2019; Sakuraba et al., 2020; Yang et al., 2020; Yan et al., 2021), which provides a molecular strategy to improve crop yield or quality by finely regulating the leaf senescence process.
The basic helix-loop-helix (bHLH) family TFs also regulate leaf senescence. Members of bHLH subgroup IIIe factors, including myelocytomatosis protein 2 (MYC2), MYC3 and MYC4, antagonistically interact with the bHLH subgroup IIId factors bHLH03 (JAM3), bHLH13 (JAM2), bHLH14, and bHLH17 (JAM1), and mediate JA-induced leaf senescence by directly binding the promoter of SAG29 in Arabidopsis (Qi et al., 2015; Goossens et al., 2017). In addition, MYC5 also positively regulates JA-induced leaf senescence (Song et al., 2017). Darkness induces the protein accumulation of JASMONATE ZIM-domain 7(JAZ7), which in turn inhibits dark-induced leaf senescence by suppressing MYC2 (Yu et al., 2016). In rice, OsMYC2 acts as a positive regulator of leaf senescence by regulating the transcript levels of SAGs (Uji et al., 2017). These findings suggest that MYC2 regulates leaf senescence via multiple signaling pathways. ACTIVATION-TAGGED BRI1 (BRASSINOSTEROID-INSENSITIVE1)-SUPPRESSOR1 (ATBS1)-INTERACTING FACTOR2 (AIF2) is a non-DNA-binding bHLH TF and delays dark or BR-induced leaf senescence (Kim et al., 2020). Phytochrome-interacting bHLH transcription factors (PIFs) such as PIF4 and PIF5 promote leaf senescence under natural or dark conditions in Arabidopsis (Sakuraba et al., 2014; Song et al., 2014; Li N. et al., 2021). MdbHLH3 regulates leaf senescence by promoting the expression of dehydratase-enolase-phosphatase complex 1 (MdDEP1) in Malus Domestica (Hu et al., 2020). These results imply that bHLH TFs are involved in the regulation of leaf senescence in both annuals and perennial woody plants.
There is growing evidence that multiple TF families of genes are involved in the regulation of leaf senescence, including MYB-TFs such as MYB2 (Buchanan-Wollaston et al., 2005), MYB DOMAIN PROTEIN R1 (MYBR1) (Jaradat et al., 2013), MYB HYPOCOTYL ELONGATION-RELATED (MYBH) (Huang et al., 2015), AtMYBL (Zhang et al., 2011), OsMYB102 (Piao et al., 2019), ScMYB2S1 (Guo J. et al., 2017); PLANT A/T-RICH SEQUENCE- AND ZINC-BINDING PROTEIN (PLATZ) family transcription factor (ORE15) (Kim J. H. et al., 2018); AP2/ERF transcription factors such as CRF1/2/3/5/6 in Arabidopsis (Raines et al., 2016), SlERF36 in Tomato (Upadhyay et al., 2013), and SUBMERGENCE1A (SUB1A) in Rice (Fukao et al., 2012); AP2/DREB transcription factors (DEAR1 and Rap2.4f) (Tsutsui et al., 2009; Xu et al., 2010); CCCH zinc-finger family [K-homolog (KH) proteins, KHZ1 and KHZ2] (Yan et al., 2017); AUXIN RESISTANT 3 (AXR3)/INDOLE-3-ACETIC ACID INDUCIBLE 17 (IAA17), one member of Auxin response factors (ARF) family, is a positive regulator of natural leaf senescence (Shi et al., 2015); TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) family transcription factor (TCP2/4/10) (Schommer et al., 2008); Homeodomain-leucine zipper family (REVOLUTA) (Xie Y. et al., 2014); MADS box transcription factors such as FOREVER YOUNG FLOWER (FYF) in Arabidopsis and SlFYFL in Tomato (Chen et al., 2011; Xie Q. et al., 2014; Guo X. et al., 2017), as well as RAV family transcription factor (RAV1) (Woo et al., 2010).
A large number of studies have shown that TF plays a key regulatory role in leaf senescence, however, most of the studies have mainly focused on a few families, including the NAC or WRKY families, and more studies are needed in the future to analyze whether other family TFs are also involved in leaf senescence. A recent transcriptomic study revealed that 115 Sen-TFs from 31 families are involved in autumn leaf senescence in poplar (Wang et al., 2021), further supporting this suggestion.
Post-transcriptional Level
Post-transcriptional regulation, including RNA editing, polyadenylation, mRNA stability, and alternative splicing, is related to leaf senescence. Multiple organellar RNA editing factors 9 (MORF9), one of the core proteins of plant editosomes, are involved in the RNA editing in chloroplasts, and its mRNA level declined in senescent leaves (Tian et al., 2019). MicroRNA (miRNA) is involved in leaf senescence by regulating the expression of SAG genes. For example, miR156, miR164, miR172, and miR840 regulate leaf senescence by suppressing their target genes (Kim et al., 2009; Wu et al., 2016, 2020; Tian et al., 2019; Yujun et al., 2019; Roussin-Leveillee et al., 2020). In addition to miRNAs, circular RNA (circRNA), and long non-coding RNA (lncRNAs) participate in leaf senescence of rice by a competitive endogenous RNA (CeRNA) network (Huang et al., 2021a, b). Interestingly, EIN3 and clock-associated PSEUDO-RESPONSE REGULATOR 9 (PRR9) up-regulate the transcription level of ORE1 by inhibiting the transcription of miR164 (Li et al., 2013; Kim H. et al., 2018). miR398 participates in regulating leaf senescence by post-transcriptional regulation of ASCORBATE PEROXIDASE 6 (APX6) (Chen et al., 2021). SlymiR208 regulates leaf senescence by controlling the expression of isopentenyl transferases SlIPT2 and SlIPT4 (Zhang Y. et al., 2020).
Alternative splicing (AS) is widely used in RNA splicing and processing after gene transcription in higher eukaryotes, which can increase the diversity of transcriptome and proteome. AS events can be mainly classified into five categories: IR, skipping exon and mutually exclusive exons, as well as alternative 5’-splice sites and alternative 3’-splice sites. In animals, splicing factors control cellular senescence by regulating the splicing process of RNA precursors (Fregoso et al., 2013). In plants, AS acts as a regulatory mechanism of plant development or adaptation to environmental stress factors. RNA splicing factor RNA-BINDING PROTEIN 25 (RBM25) responds to ABA stress in Arabidopsis (Zhan et al., 2015). Loss-of-function of CRM FAMILY MEMBER SUBFAMILY 4 (CFM4) leads to abnormal rRNA processing during chloroplast RNA splicing, and exhibited plant growth retardation and delayed senescence (Lee et al., 2014). An interesting discovery shows that the differential expression of sugarcane MYB TF ScMYB2 alternative splicing transcripts may be an important post-transcriptional regulatory mechanism for controlling drought stress and leaf senescence (Guo J. et al., 2017).
The splicing mechanism occurs in the spliceosome, which is composed of five small nuclear RNAs (snRNAs) and a series of related protein factors. The spliceosome can recognize the splice site of the precursor RNA and catalyze the splicing reaction. There are major spliceosomes (U2) and minor spliceosomes (U12) that support splicing functions (Sharp, 2005). Although the splicing efficiency of the U12-type spliceosome is relatively lower, splicing errors will affect the normal growth and development of plants. The U12-type intron-specific small spliceosome mainly removes the small U12 intron from the precursor mRNA. Mutation of u11-48k causes defects in growth and development, such as short plant size, increased lotus-like leaves, and delayed senescence (Xu et al., 2016), indicating that the regulation of the RNA splicing process has a potentially important effect on plant leaf senescence. ETHYLENE RESPONSE FACTOR4 (ERF4) has two different isoforms, ERF4-R and ERF4-A, produced by alternative polyadenylation of its pre-mRNA. ERF4-R, contains an ERF-associated amphiphilic repression (EAR) motif and acts as a repressor, whereas the other form, ERF4-A, is lacking this motif and acts as an activator. ERF4-R and ERF4-A can directly bind to the promoter of CATALASE3 (CAT3) but have antagonistic effects on gene expression. The ratio of ERF4-A to ERF4-R mRNA changed as the plant ages and caused a complex age-dependent regulation of CAT3 activity. Interestingly, overexpression of ERF4-R but not of ERF4-A led to accelerated senescence (Koyama et al., 2013; Riester et al., 2019). ONAC054 was shown to participate in ABA-induced leaf senescence by directly activating OsABI5 in rice (Sakuraba et al., 2020). Interestingly, the ONAC054 transcript (ONAC054α) has an alternatively spliced form, ONAC054β, encoding a small truncated protein. Overexpression of ONAC054α or ONAC054β promotes leaf senescence (Sakuraba et al., 2020). A recent study reported that an alternative splicing event retaining the first intron of the PtRD26 pre-mRNA occurred in a senescence-associated manner in poplar. The intron retention (IR) event in PtRD26 led to an alternative splicing variant, PtRD26IR, which encodes a truncated protein. PtRD26IR forms heterodimers with multiple hub Sen-NAC TFs, including PtNAC039, PtNAC055, PtNAC076, PtNAC086, PtNAC099, and PtNAC109, represses their DNA binding activity to target genes, and delays age-, dark,- and PtRD26-induced leaf senescence in poplar, tobacco, and Arabidopsis. PtRD26 regulates Sen-NAC TFs by directly binding their promoters or indirectly through protein-protein interactions using its splicing variant, PtRD26IR, thereby forming a multiply-interlocked feed-forward loop to finely tune the leaf senescence process. Functional analysis of senescence-associated splicing factors (SF) revealed that PtU2A2A, PtU2A2B-1, or PtU2A2B-2 (U2 auxiliary factor large subunit A or B) are involved in AS of PtRD26IR. Silencing separately or simultaneously of these SFs significantly decreased the transcript levels of PtRD26IR and accelerated leaf senescence. Based on these findings, it is found that the products of AS have different functions and regulate plant development such as plant senescence through different mechanisms. With the application of multi-omics technology, more AS events will be found to be involved in the regulation of leaf senescence, which will Further deepen the mechanistic understanding of plant aging.
Translation Level
Senescence is a long-term state of cell cycle arrest arising from cells that have suffered sublethal damage. Although senescent cells no longer replicate, they remain metabolically active and further develop a distinct and stable phenotype not seen in proliferating cells (Guo and Gan, 2005; Lim et al., 2007). On the one hand, along with leaf senescence, a large number of proteins are degraded and translation efficiency decreases; on the other hand, senescence-specific regulatory factors are synthesized to inhibit or retard the leaf senescence process (Guo and Gan, 2005; Lim et al., 2007). Thus, translation in senescent cells paradoxically includes a general inhibition of translation triggered by numerous stresses and a selective increase in translation of specific proteins, including SAG protein.
Mutation of ORE4, which encodes the plastid ribosomal small subunit protein 17 that is a component of the plastid ribosome, reduces the translation rate in the chloroplast and thus extends leaf longevity in Arabidopsis (Woo et al., 2002), suggesting a possible link between decreased metabolism and extended longevity of the leaves. Translation initiation, the first step in the protein synthesis process, is the main regulatory step controlling translation and involves a large number of translation initiation factors. Studies in plants have revealed that these translation initiation factors affect various aspects of plant growth and development, in addition to their role in protein synthesis (Wang et al., 2001). Mutation of EUKARYOTIC ELONGATION FACTOR 5A (eIF5A) significantly inhibits plant nutrition and reproductive growth and delays leaf senescence in Arabidopsis and Picrorhiza (Picrorhiza kurrooa Royle ex Benth.) (Reviron et al., 1992; Parkash et al., 2014). Translation initiation factor eIF3h is involved in the signal activation and restart of rapamycin (TOR) and affects the growth and development of plants (Schepetilnikov et al., 2013). In addition, eukaryotic translation elongation factors (eEF) also involved leaf senescence. For instance, mutation of Spotted Leaf 33 (spl33), encoding a eEF1 alpha (eEF1A)-like protein, induces early leaf senescence (Wang et al., 2017). Ectopic expression of Lilium regales Eukaryotic translation elongation factor 1 alpha 4 (LreEF1A4), encoding the α subunit of elongation factor 1 from a Lilium regale cucumber mosaic virus (CMV), delayed leaf and flower senescence in petunia (Petunia hybrida) (Sun et al., 2020). Interestingly, two subunits of ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco), the key enzyme that determines the rate of carbon assimilation in photosynthesis, is controllable at the translation level, and affect plant growth and development, including the leaf senescence process (Suzuki and Makino, 2013; Woo et al., 2013).
Post-translational Level
Post-translational modifications (PTM), including methylation, acetylation, phosphorylation, ubiquitination, and deubiquitination affect the structure and function of proteins. Previous studies found that the PTM of a large number of SAG proteins changed with leaf senescence (Wang and Schippers, 2019). This implies a close relationship between leaf senescence and PTM, but the causal relationship is not clear.
Transcriptomics analysis reveals that a large number of SAGs are involved in PTM, such as receptor-like kinase (RLK) and mitogen-activated protein kinase (MAPK) (Ahmad and Guo, 2019). RLK is an ideal candidate for senescence-inducing signal receptors, which often have an N-terminal extracellular binding domain for ligand binding, a transmembrane domain spanning the plasma membrane, and a cytoplasmic kinase domain (Shiu and Bleecker, 2001; Gish and Clark, 2011). The largest subfamily of RLK is the leucine-rich repeat receptor-like protein kinase (LRR-RLK), containing more than 200 members, and lots of them are involved in the regulation of leaf senescence (Shiu et al., 2004). GmSARK (Glycine max Senescence-Associated Receptor-like Kinase), a senescence-associated LRR-RLK isolated from soybean (Glycine max) and its homolog AtSARK in Arabidopsis are positive regulators of leaf senescence (Li et al., 2006; Xu et al., 2011). SARK-mediated signaling pathway positively regulates leaf senescence through suppressing SMALL AUXIN-UP RNA 49 (SAUR49), a negative regulator of leaf senescence, and activating SENESCENCE-SUPPRESSED PROTEIN PHOSPHATASE (SSPP), an accelerator of leaf senescence (Xiao et al., 2015; Wen et al., 2020). In contrast, the somatic embryogenesis receptor-like kinase 4 (SERK4) and the cell wall-associated kinase 10 (AtWAKL10) act as the negative regulators of leaf senescence (Li et al., 2019; Li L. et al., 2021). Interestingly, a common receptor can work with multiple receptors in different signaling pathways. AtSARK and SERK4 may be part of the receptor complex that regulates plant aging by acting with other LRR-RLKs (Brandt and Hothorn, 2016; Cui et al., 2018).
The mitogen-activated protein kinase cascade MAPKKK-MAPKK-MAPK is one of the most important signal transduction pathways in plants and animals. Recently, MAP KINASE 4/5 (MKK4/5)-MITOGEN-ACTIVATED PROTEIN KINASE 1/2 (MPK1/2), MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 18 (MAPKKK18), and OsMAPKKK1 have been found to be the positive regulators of leaf senescence (Matsuoka et al., 2015; Wang et al., 2015; Zhang J. et al., 2020). By contrast, Enhanced Disease Resistance 1 (EDR1), a MAPKK, functions as a negative regulator by coordinating biotic stress response and ethylene-induced senescence (Frye et al., 2001; Tang and Innes, 2002). MKK9 phosphorylates the target MPK6, which stabilizes the leaf senescence transcription factor EIN3 by promoting the cleavage and nuclear translocation of ORE3/EIN2 (Zhou et al., 2009; Zhang Y. et al., 2016). These findings suggest that RLKs and MAPKs regulate leaf senescence by affecting the phosphorylation status of target proteins.
The leaf senescence process is accompanied by protein degradation. The main protein degradation pathways are autophagy and the ubiquitin-proteasome system (UPS), which precisely regulate the turnover of organelles and the degradation of abnormal proteins and maintain protein homeostasis. Autophagy and protein ubiquitination are synergistic in the cell. Ubiquitination acts as a signal to induce organelles to target autophagy. Mitophagy and chloroplast protein degradation is the result of the synergistic effect of ubiquitination and autophagy (Geisler et al., 2010; Kikuchi et al., 2020). Interestingly, autophagy seems to prevent aging, whereas the proteasome acts as a positive regulator of aging (Wang and Schippers, 2019). Chaperone-mediated autophagy is one of the main types of autophagy in cells, with high selectivity. Autophagy-related genes (ATG) involved in autophagy are up-regulated with the occurrence of plant senescence (Masclaux-Daubresse et al., 2014). Mutation of several ATG genes, including ATG4a/4b, ATG9, ATG19, and ATG18a, promotes leaf senescence under nitrogen-starvation conditions (Hanaoka et al., 2002; Yoshimoto et al., 2004; Xiong et al., 2005; Phillips et al., 2008; Wang and Schippers, 2019). Although most studies support the role of autophagy in delaying aging, ATG8 promotes senescence by interacting with the ABNORMAL SHOOT3 (ABS3). This non-autophagic ATG8-ABS3 pathway interacts with the classic autophagy pathway to balance aging and survival (Jia et al., 2019). Therefore, the components of autophagy may have a dual role in the initiation and progression of senescence. 26S proteasome is mainly responsible for degrading ubiquitinated proteins. The recognition of ubiquitinated substrates in the process of ubiquitin/proteasome-mediated proteolysis (UPP) is directly mediated by the proteasome subunits RPN10 (REGULATORY PARTICLE NON-ATPase 10) and RPN13. The loss of the potential UPP ubiquitin receptor RPN10 significantly delays senescence (Lin et al., 2011), and overexpression of RPN5a leads to premature senescence (Book et al., 2009). In contrast to the overall up-regulation of ATG genes, transcript levels of only a small part of the proteasome subunit genes were increased during leaf senescence (Guo and Gan, 2012). In the senescent leaf of rape and barley (Hordeum vulgare L.), the proteasome is very active (Poret et al., 2016; Velasco-Arroyo et al., 2016). Interestingly, an application of protease inhibitor delays the onset of senescence symptoms (Pak and van Doorn, 2005). Taken together, these observations imply that autophagy and proteasome seem to have different effects on the onset of senescence, and they coordinately regulate the progression of leaf senescence.
One of the well-characterized PTMs involved in the regulation of leaf senescence is ubiquitination/deubiquitination modification. Protein ubiquitination requires the synergy of ubiquitin activation (E1), ubiquitin-binding (E2), and ubiquitin ligase (E3). Members of E2 and E3 have been found to be involved in the regulation of leaf senescence (Shu and Yang, 2017; Park et al., 2018). Among them, RING-type E3 and U-box-type E3 ligases have been shown to act as regulators of leaf senescence by mediating ABA signaling. For example, PLANT U-box (PUB) E3 ubiquitin ligase PUB12 and PUB13 ubiquitinated FLS2 (FLAGELLIN-SENSITIVE 2) for protein degradation, thereby down-regulating flagellin signaling and negatively regulating stress-induced leaf senescence (Zhou et al., 2015). In addition, HECT-type ubiquitin E3 ligase (UPL1-UPL7) plays a critical role in cell death and leaf senescence (Lan and Miao, 2019). Mutation of UBIQUITIN PROTEIN LIGASE 5 (UPL5) leads to the accumulation of WRKY53 and induces early leaf senescence (Zentgraf et al., 2010). Ubiquitin-specific protease (UBP1)-associated protein 2a (UBA2a), UBA2b, and UBA2c positive regulators of leaf senescence (Kim et al., 2008). Likewise, the potato (Solanum tuberosum) RNA-binding protein StUBA2a/b is homologous to Arabidopsis UBA2s. Constitutive overexpression of StUBA2a/b increases the expression of the SAG13 gene, pathogen-related genes (PR), and autophagy-related genes, and promotes leaf senescence in Arabidopsis (Na et al., 2015). The process of protein ubiquitination is reversible, and deubiquitinating enzymes (DUBs) can remove mono-ubiquitin molecules or polyubiquitin chains on proteins. UBP is the largest DUB subfamily, and members of the UBP family are involved in a variety of physiological processes, including leaf senescence (Zhou et al., 2017). Out of them, UBIQUITIN-SPECIFIC PROTEASE 12 (UBP12) and UBP13 are involved in the regulation of circadian clock and flowering (Cui et al., 2013), and accelerate nitrogen starvation-induced leaf senescence by counteracting the effect of E3 ligase NLA (Nitrogen Ubiquitin-Protein Ligases DNA) to maintain the homeostasis of ORE1 (Park et al., 2019).
Conclusions and Perspectives
Leaf senescence is a highly complex process of orderly degradation of cell structure and is controlled by multiple layers regulatory network (Figure 1), in which different regulatory factors at different levels may interact to fine-tune the initiation and progression of leaf senescence (Table 1). Although regulation is artificially divided into multiple levels (Woo et al., 2013), leaf senescence is a highly dynamic regulatory process (Woo et al., 2019), and there is no single way to regulate it. For example, changes in chromatin structure affect gene expression, protein translation, and thus the function of transcription factors, which in turn cause changes in the senescence process of plant leaves. Moreover, the regulation of leaf senescence involves not only the interactions between proteins, proteins, and DNA, but also the exchange of information between cells and organelles, thus synergistically regulating the initiation of leaf senescence, which guarantees the return of nutrients and the survival of plants. Therefore, we should combine genome, transcriptome, proteome, metabolome, and the latest translation comics data to discuss the general mechanism of regulate senescence and understand how senescence and death are systemically integrated within the entire plant (Kim J. et al., 2016).
With the aid of forwarding or reversing genetics strategies and the development of multi-functional CRISPR genome editing technology, a large number of senescence-related mutants will be generated. For example, quintuple mutants of oss40s-cr generated using CRISPR technology displays stay-green phenotypes (Habiba et al., 2021), which will further deepen our understanding of leaf senescence. The model plant Arabidopsis has played an important role in revealing the molecular or genetic regulation mechanisms of plant senescence, but we still know little about leaf senescence and do not fully understand the biological significance of senescence (Lim et al., 2007). The relatively short life cycle of Arabidopsis has limitations for our understanding of plant aging. Along with the genomic information revealed for a variety of plants, it provides the possibility to systematically study plant senescence by comparative genomics.
It’s unclear how these transcription factors regulate, such as the WRKY family and NAC family, and epigenetic factors co-regulate the senescence process of plants. The function of hormone signaling on leaf senescence has been widely recognized (Hu et al., 2017). It is necessary to further explore how plant signals and environmental signals are integrated into the hormone signaling pathway, and how post-translational modifications such as phosphorylation and ubiquitination are passed through transcription factors, kinases, and protease, finely control these signals to regulate gene expression and protein turnover during leaf senescence. The senescence symptoms of leaf senescence have always been detected at the organ level. However, in senescent leaves, leaf cells are usually at different developmental ages or senescence stages, which makes it impossible to better understand the biological process of leaf senescence. Fortunately, the application of single-cell sequencing technology may offer the possibility to resolve the cytological basis of leaf senescence.
In addition to the loss- or gain-of-function of mutants, ecotypes of various species will greatly contribute to the understanding of the molecular mechanisms underlying leaf senescence. Through analysis of naturally occurring DNA methylation variation regions (NMRs) between Col-0 and C24 accessions of Arabidopsis thaliana, a retrotransposon named NMR19-4 (naturally occurring DNA methylation variation region 19) was identified to be involved in the regulation of leaf senescence (He et al., 2018). NMR19-4 is an environmentally associated epiallele that controls leaf senescence by regulating the expression of PHEOPHYTIN PHEOPHORBIDE HYDROLASE (PPH), which is involved in chlorophyll breakdown (Schelbert et al., 2009; He et al., 2018). By mapping the quantitative trait locus (QTL) of leaf senescence between the Col-0 and Ct-1 accessions of Arabidopsis thaliana, ACCELERATED CELL DEATH 6 (ACD6) was identified as the causal gene (Jasinski et al., 2020). Using two rice subspecies indica and japonica, variations were found in the promoter regions of the Stay-Green (OsSGR) gene encoding a chlorophyll-degrading enzyme. This promoter variations trigger higher and earlier induction of OsSGR, which in turn accelerates leaf senescence in indica (Shin et al., 2020).
Author Contributions
ZL conceived the project and designed the manuscript. HG and XX designed part of the manuscript. Y-MZ collected the data and organized figure. PG organized table. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely apologize to those authors whose work is not included in this review due to space limitations. We thank Hou-Ling Wang (Beijing Forestry University) for figure preparation.
Funding
This work was funded by grants from the National Natural Science Foundation of China (Nos. 32170345, 31970196, and 32011540381 to ZL), the National Key Research and Development Program of China (No. 2019YFA0903904 to HG), the Shenzhen Science and Technology Program (No. KQTD20190929173906742 to HG), and the startup funding for plant aging research from Beijing Forestry University (No. BJFU2021YJRC00600K).
References
- Ahmad S., Guo Y. (2019). Signal Transduction in Leaf Senescence: Progress and Perspective. Plants 8:10. 10.3390/plants8100405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archacki R., Yatusevich R., Buszewicz D., Krzyczmonik K., Patryn J., Iwanicka-Nowicka R., et al. (2017). Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucleic Acids Res. 45 3116–3129. 10.1093/nar/gkw1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay N., Irmler K., Fischer A., Uhlemann R., Reuter G., Humbeck K. (2009). Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 58 333–346. 10.1111/j.1365-313X.2008.03782.x [DOI] [PubMed] [Google Scholar]
- Balanza V., Martinez-Fernandez I., Sato S., Yanofsky M. F., Kaufmann K., Angenent G. C., et al. (2018). Genetic control of meristem arrest and life span in Arabidopsis by a FRUITFULL-APETALA2 pathway. Nat. Commun. 9:565. 10.1038/s41467-018-03067-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S., Kwasniewski M., Caldana C., Mehrnia M., Zanor M. I., Xue G. P., et al. (2011). ORS1, an H(2)O(2)-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 4 346–360. 10.1093/mp/ssq080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J., Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21 381–395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S., Li J., Palva E. T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63 2667–2679. 10.1093/jxb/err450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Book A. J., Smalle J., Lee K. H., Yang P., Walker J. M., Casper S., et al. (2009). The RPN5 subunit of the 26s proteasome is essential for gametogenesis, sporophyte development, and complex assembly in Arabidopsis. Plant Cell 21 460–478. 10.1105/tpc.108.064444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B., Hothorn M. (2016). SERK co-receptor kinases. Curr. Biol. 26 R225–R226. 10.1016/j.cub.2015.12.014 [DOI] [PubMed] [Google Scholar]
- Breeze E., Harrison E., McHattie S., Hughes L., Hickman R., Hill C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23 873–894. 10.1105/tpc.111.083345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusslan J. A., Bonora G., Rus-Canterbury A. M., Tariq F., Jaroszewicz A., Pellegrini M. (2015). A Genome-Wide Chronological Study of Gene Expression and Two Histone Modifications, H3K4me3 and H3K9ac, during Developmental Leaf Senescence. Plant Physiol. 168 1246–1261. 10.1104/pp.114.252999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusslan J. A., Rus Alvarez-Canterbury A. M., Nair N. U., Rice J. C., Hitchler M. J., Pellegrini M. (2012). Genome-wide evaluation of histone methylation changes associated with leaf senescence in Arabidopsis. PLoS One 7:e33151. 10.1371/journal.pone.0033151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P. O., Nam H. G., et al. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42 567–585. 10.1111/j.1365-313X.2005.02399.x [DOI] [PubMed] [Google Scholar]
- Buszewicz D., Archacki R., Palusinski A., Kotlinski M., Fogtman A., Iwanicka-Nowicka R., et al. (2016). HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 39 2108–2122. 10.1111/pce.12756 [DOI] [PubMed] [Google Scholar]
- Chen C., Galon Y., Rahmati Ishka M., Malihi S., Shimanovsky V., Twito S., et al. (2021). ASCORBATE PEROXIDASE6 delays the onset of age-dependent leaf senescence. Plant Physiol. 185 441–456. 10.1093/plphys/kiaa031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiang S., Chen Y., Li D., Yu D. (2017). Arabidopsis WRKY45 Interacts with the DELLA Protein RGL1 to Positively Regulate Age-Triggered Leaf Senescence. Mol. Plant 10 1174–1189. 10.1016/j.molp.2017.07.008 [DOI] [PubMed] [Google Scholar]
- Chen L. J., Wuriyanghan H., Zhang Y. Q., Duan K. X., Chen H. W., Li Q. T., et al. (2013). An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol. 163 1752–1765. 10.1104/pp.113.224881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. K., Hsu W. H., Lee P. F., Thiruvengadam M., Chen H. I., Yang C. H. (2011). The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J. 68 168–185. 10.1111/j.1365-313X.2011.04677.x [DOI] [PubMed] [Google Scholar]
- Chen S. P., Lin I. W., Chen X., Huang Y. H., Chang S. C., Lo H. S., et al. (2016). Sweet potato NAC transcription factor, IbNAC1, upregulates sporamin gene expression by binding the SWRE motif against mechanical wounding and herbivore attack. Plant J. 86 234–248. 10.1111/tpj.13171 [DOI] [PubMed] [Google Scholar]
- Cho E. J., Choi S. H., Kim J. H., Kim J. E., Lee M. H., Chung B. Y., et al. (2016). A Mutation in Plant-Specific SWI2/SNF2-Like Chromatin-Remodeling Proteins, DRD1 and DDM1, Delays Leaf Senescence in Arabidopsis thaliana. PLoS One 11:e0146826. 10.1371/journal.pone.0146826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Lu F., Li Y., Xue Y., Kang Y., Zhang S., et al. (2013). Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol. 162 897–906. 10.1104/pp.112.213009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Zhao P., Liang W., Cheng Q., Mu B., Niu F., et al. (2020). A Rapeseed WRKY Transcription Factor Phosphorylated by CPK Modulates Cell Death and Leaf Senescence by Regulating the Expression of ROS and SA-Synthesis-Related Genes. J. Agric. Food Chem. 68 7348–7359. 10.1021/acs.jafc.0c02500 [DOI] [PubMed] [Google Scholar]
- Cui Y., Hu C., Zhu Y., Cheng K., Li X., Wei Z., et al. (2018). CIK Receptor Kinases Determine Cell Fate Specification during Early Anther Development in Arabidopsis. Plant Cell 30 2383–2401. 10.1105/tpc.17.00586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey C. A., Sargent D. F., Luger K., Maeder A. W., Richmond T. J. (2002). Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 319 1097–1113. 10.1016/S0022-2836(02)00386-8 [DOI] [PubMed] [Google Scholar]
- Efroni I., Han S. K., Kim H. J., Wu M. F., Steiner E., Birnbaum K. D., et al. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24 438–445. 10.1016/j.devcel.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mannai Y., Akabane K., Hiratsu K., Satoh-Nagasawa N., Wabiko H. (2017). The NAC Transcription Factor Gene OsY37 (ONAC011) Promotes Leaf Senescence and Accelerates Heading Time in Rice. Int. J. Mol. Sci. 18:10. 10.3390/ijms18102165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z. Q., Tan X. L., Shan W., Kuang J. F., Lu W. J., Chen J. Y. (2018). Characterization of a Transcriptional Regulator, BrWRKY6, Associated with Gibberellin-Suppressed Leaf Senescence of Chinese Flowering Cabbage. J. Agric Food Chem. 66 1791–1799. 10.1021/acs.jafc.7b06085 [DOI] [PubMed] [Google Scholar]
- Fregoso O. I., Das S., Akerman M., Krainer A. R. (2013). Splicing-factor oncoprotein SRSF1 stabilizes p53 via RPL5 and induces cellular senescence. Mol. Cell 50 56–66. 10.1016/j.molcel.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye C. A., Tang D., Innes R. W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. U S A 98 373–378. 10.1073/pnas.011405198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T., Yeung E., Bailey-Serres J. (2012). The submergence tolerance gene SUB1A delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiol. 160 1795–1807. 10.1104/pp.112.207738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapati P., Xue G. P., Munne-Bosch S., Balazadeh S. (2015). Transcription Factor ATAF1 in Arabidopsis Promotes Senescence by Direct Regulation of Key Chloroplast Maintenance and Senescence Transcriptional Cascades. Plant Physiol. 168 1122–1139. 10.1104/pp.15.00567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S., Holmstrom K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., et al. (2010). PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12 119–131. 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- Gish L. A., Clark S. E. (2011). The RLK/Pelle family of kinases. Plant J. 66 117–127. 10.1111/j.1365-313X.2011.04518.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens J., Mertens J., Goossens A. (2017). Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 68 1333–1347. 10.1093/jxb/erw440 [DOI] [PubMed] [Google Scholar]
- Gu L., Ma Q., Zhang C., Wang C., Wei H., Wang H., et al. (2019). The Cotton GhWRKY91 Transcription Factor Mediates Leaf Senescence and Responses to Drought Stress in Transgenic Arabidopsis thaliana. Front. Plant Sci. 10:1352. 10.3389/fpls.2019.01352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Wei H., Wang H., Su J., Yu S. (2018). Characterization and functional analysis of GhWRKY42, a group IId WRKY gene, in upland cotton (Gossypium hirsutum L.). BMC Genet. 19:48. 10.1186/s12863-018-0653-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Ling H., Ma J., Chen Y., Su Y., Lin Q., et al. (2017). A sugarcane R2R3-MYB transcription factor gene is alternatively spliced during drought stress. Sci. Rep. 7:41922. 10.1038/srep41922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P., Li Z., Huang P., Li B., Fang S., Chu J., et al. (2017). A Tripartite Amplification Loop Involving the Transcription Factor WRKY75, Salicylic Acid, and Reactive Oxygen Species Accelerates Leaf Senescence. Plant Cell 29 2854–2870. 10.1105/tpc.17.00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Chen G., Naeem M., Yu X., Tang B., Li A., et al. (2017). The MADS-box gene SlMBP11 regulates plant architecture and affects reproductive development in tomato plants. Plant Sci. 258 90–101. 10.1016/j.plantsci.2017.02.005 [DOI] [PubMed] [Google Scholar]
- Guo Y., Gan S. (2005). Leaf senescence: signals, execution, and regulation. Curr. Top. Dev. Biol. 71 83–112. 10.1016/S0070-2153(05)71003-6 [DOI] [PubMed] [Google Scholar]
- Guo Y., Gan S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46 601–612. 10.1111/j.1365-313X.2006.02723.x [DOI] [PubMed] [Google Scholar]
- Guo Y., Gan S. S. (2012). Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 35 644–655. 10.1111/j.1365-3040.2011.02442.x [DOI] [PubMed] [Google Scholar]
- Habiba, Xu J., Gad A. G., Luo Y., Fan C., Uddin J. B. G., et al. (2021). Five OsS40 Family Members Are Identified as Senescence-Related Genes in Rice by Reverse Genetics Approach. Front Plant Sci. 12:701529. 10.3389/fpls.2021.701529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Du M., Zhou Z., Wang S., Li T., Han J., et al. (2020). Overexpression of a Malus baccata NAC Transcription Factor Gene MbNAC25 Increases Cold and Salinity Tolerance in Arabidopsis. Int. J. Mol. Sci. 21:4. 10.3390/ijms21041198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Kim C. Y., Lee J., Lee S. K., Jeon J. S. (2014). OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol. Cells 37 532–539. 10.14348/molcells.2014.0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., et al. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129 1181–1193. 10.1104/pp.011024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Wu W., Zinta G., Yang L., Wang D., Liu R., et al. (2018). A naturally occurring epiallele associates with leaf senescence and local climate adaptation in Arabidopsis accessions. Nat. Commun. 9:460. 10.1038/s41467-018-02839-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman R., Hill C., Penfold C. A., Breeze E., Bowden L., Moore J. D., et al. (2013). A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 75 26–39. 10.1111/tpj.12194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinckley W. E., Keymanesh K., Cordova J. A., Brusslan J. A. (2019). The HAC1 histone acetyltransferase promotes leaf senescence and regulates the expression of ERF022. Plant Direct. 3:e00159. 10.1002/pld3.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D. G., Sun C. H., Zhang Q. Y., Gu K. D., Hao Y. J. (2020). The basic helix-loop-helix transcription factor MdbHLH3 modulates leaf senescence in apple via the regulation of dehydratase-enolase-phosphatase complex 1. Hortic Res. 7:50. 10.1038/s41438-020-0273-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Jiang Y., Han X., Wang H., Pan J., Yu D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68 1361–1369. 10.1093/jxb/erx004 [DOI] [PubMed] [Google Scholar]
- Hu Y., Lu Y., Zhao Y., Zhou D. X. (2019). Histone Acetylation Dynamics Integrates Metabolic Activity to Regulate Plant Response to Stress. Front. Plant Sci. 10:1236. 10.3389/fpls.2019.01236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. K., Lo P. C., Huang L. F., Wu S. J., Yeh C. H., Lu C. A. (2015). A single-repeat MYB transcription repressor, MYBH, participates in regulation of leaf senescence in Arabidopsis. Plant Mol. Biol. 88 269–286. 10.1007/s11103-015-0321-2 [DOI] [PubMed] [Google Scholar]
- Huang R., Liu D., Huang M., Ma J., Li Z., Li M., et al. (2019). CpWRKY71, a WRKY Transcription Factor Gene of Wintersweet (Chimonanthus praecox), Promotes Flowering and Leaf Senescence in Arabidopsis. Int. J. Mol. Sci. 20:21. 10.3390/ijms20215325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Zhang H., Guo R., Wang Q., Liu X., Kuang W., et al. (2021a). Systematic identification and characterization of circular RNAs involved in flag leaf senescence of rice. Planta 253:26. 10.1007/s00425-020-03544-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Zhang H., Wang Q., Guo R., Wei L., Song H., et al. (2021b). Genome-wide identification and characterization of long non-coding RNAs involved in flag leaf senescence of rice. Plant Mol. Biol. 105 655–684. 10.1007/s11103-021-01121-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaradat M. R., Feurtado J. A., Huang D., Lu Y., Cutler A. J. (2013). Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 13:192. 10.1186/1471-2229-13-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S., Fabrissin I., Masson A., Marmagne A., Lecureuil A., Bill L., et al. (2020). ACCELERATED CELL DEATH 6 Acts on Natural Leaf Senescence and Nitrogen Fluxes in Arabidopsis. Front. Plant Sci. 11:611170. 10.3389/fpls.2020.611170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia M., Liu X., Xue H., Wu Y., Shi L., Wang R., et al. (2019). Noncanonical ATG8-ABS3 interaction controls senescence in plants. Nat. Plants 5 212–224. 10.1038/s41477-018-0348-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yang S., Yu D. (2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26 230–245. 10.1105/tpc.113.117838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H. C., Sturre M. J., Hille J., Dijkwel P. P. (2002). Arabidopsis onset of leaf death mutants identify a regulatory pathway controlling leaf senescence. Plant J. 32 51–63. 10.1046/j.1365-313x.2002.01400.x [DOI] [PubMed] [Google Scholar]
- Kan C., Zhang Y., Wang H. L., Shen Y., Xia X., Guo H., et al. (2021). Transcription Factor NAC075 Delays Leaf Senescence by Deterring Reactive Oxygen Species Accumulation in Arabidopsis. Front. Plant Sci. 12:634040. 10.3389/fpls.2021.634040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K., Shim Y., Gi E., An G., Paek N. C. (2019). Mutation of ONAC096 Enhances Grain Yield by Increasing Panicle Number and Delaying Leaf Senescence during Grain Filling in Rice. Int. J. Mol. Sci. 20:20. 10.3390/ijms20205241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Nakamura S., Woodson J. D., Ishida H., Ling Q., Hidema J., et al. (2020). Chloroplast Autophagy and Ubiquitination Combine to Manage Oxidative Damage and Starvation Responses. Plant Physiol. 183 1531–1544. 10.1104/pp.20.00237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. Y., Bove J., Assmann S. M. (2008). Overexpression of wound-responsive RNA-binding proteins induces leaf senescence and hypersensitive-like cell death. New Phytol. 180 57–70. 10.1111/j.1469-8137.2008.02557.x [DOI] [PubMed] [Google Scholar]
- Kim H., Kim H. J., Vu Q. T., Jung S., McClung C. R., Hong S., et al. (2018). Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. U S A 115 8448–8453. 10.1073/pnas.1722407115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Nam H. G., Lim P. O. (2016). Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33 48–56. 10.1016/j.pbi.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Park J. H., Kim J., Kim J. J., Hong S., Kim J., et al. (2018). Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. U S A 115 E4930–E4939. 10.1073/pnas.1721523115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Woo H. R., Nam H. G. (2016). Toward Systems Understanding of Leaf Senescence: An Integrated Multi-Omics Perspective on Leaf Senescence Research. Mol. Plant 9 813–825. 10.1016/j.molp.2016.04.017 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Kim J., Jun S. E., Park S., Timilsina R., Kwon D. S., et al. (2018). ORESARA15, a PLATZ transcription factor, mediates leaf growth and senescence in Arabidopsis. New Phytol. 220 609–623. 10.1111/nph.15291 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Woo H. R., Kim J., Lim P. O., Lee I. C., Choi S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323 1053–1057. 10.1126/science.1166386 [DOI] [PubMed] [Google Scholar]
- Kim Y., Park S. U., Shin D. M., Pham G., Jeong Y. S., Kim S. H. (2020). ATBS1-INTERACTING FACTOR 2 negatively regulates dark- and brassinosteroid-induced leaf senescence through interactions with INDUCER OF CBF EXPRESSION 1. J. Exp. Bot. 71 1475–1490. 10.1093/jxb/erz533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. S., Sakuraba Y., Han S. H., Yoo S. C., Paek N. C. (2013). Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol. 54 1660–1672. 10.1093/pcp/pct113 [DOI] [PubMed] [Google Scholar]
- Kong Z., Li M., Yang W., Xu W., Xue Y. (2006). A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 141 1376–1388. 10.1104/pp.106.082941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Koyama T., Nii H., Mitsuda N., Ohta M., Kitajima S., Ohme-Takagi M., et al. (2013). A regulatory cascade involving class II ETHYLENE RESPONSE FACTOR transcriptional repressors operates in the progression of leaf senescence. Plant Physiol. 162 991–1005. 10.1104/pp.113.218115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Miao Y. (2019). New Aspects of HECT-E3 Ligases in Cell Senescence and Cell Death of Plants. Plants 8:11. 10.3390/plants8110483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. N., Lee K. H., Kim C. S. (2015). Abscisic acid receptor PYRABACTIN RESISTANCE-LIKE 8, PYL8, is involved in glucose response and dark-induced leaf senescence in Arabidopsis. Biochem. Biophys. Res. Commun. 463 24–28. 10.1016/j.bbrc.2015.05.010 [DOI] [PubMed] [Google Scholar]
- Lee K., Lee H. J., Kim D. H., Jeon Y., Pai H. S., Kang H. (2014). A nuclear-encoded chloroplast protein harboring a single CRM domain plays an important role in the Arabidopsis growth and stress response. BMC Plant Biol. 14:98. 10.1186/1471-2229-14-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Gu L., Gao L., Chen C., Wei C. Q., Qiu Q., et al. (2016). Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 48 687–693. 10.1038/ng.3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li K., Ali A., Guo Y. (2021). AtWAKL10, a Cell Wall Associated Receptor-Like Kinase, Negatively Regulates Leaf Senescence in Arabidopsis thaliana. Int. J. Mol. Sci. 22:9. 10.3390/ijms22094885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Bo C., Zhang Y., Wang L. (2021). PHYTOCHROME INTERACTING FACTORS PIF4 and PIF5 promote heat stress induced leaf senescence in Arabidopsis. J. Exp. Bot. 72 4577–4589. 10.1093/jxb/erab158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Gao J., Yao L., Ren G., Zhu X., Gao S., et al. (2016). The role of ANAC072 in the regulation of chlorophyll degradation during age- and dark-induced leaf senescence. Plant Cell Rep. 35 1729–1741. 10.1007/s00299-016-1991-1 [DOI] [PubMed] [Google Scholar]
- Li W., Li X., Chao J., Zhang Z., Wang W., Guo Y. (2018). NAC Family Transcription Factors in Tobacco and Their Potential Role in Regulating Leaf Senescence. Front. Plant Sci. 9:1900. 10.3389/fpls.2018.01900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ahmad S., Ali A., Guo C., Li H., Yu J., et al. (2019). Characterization of Somatic Embryogenesis Receptor-Like Kinase 4 as a Negative Regulator of Leaf Senescence in Arabidopsis. Cells 8:1. 10.3390/cells8010050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. P., Gan R., Li P. L., Ma Y. Y., Zhang L. W., Zhang R., et al. (2006). Identification and functional characterization of a leucine-rich repeat receptor-like kinase gene that is involved in regulation of soybean leaf senescence. Plant Mol. Biol. 61 829–844. 10.1007/s11103-006-0052-5 [DOI] [PubMed] [Google Scholar]
- Li Y., Liao S., Mei P., Pan Y., Zhang Y., Zheng X., et al. (2021). OsWRKY93 Dually Functions Between Leaf Senescence and in Response to Biotic Stress in Rice. Front. Plant Sci. 12:643011. 10.3389/fpls.2021.643011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H. (2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25 3311–3328. 10.1105/tpc.113.113340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Y., Zou D., Zhao Y., Wang H. L., Zhang Y., et al. (2020a). LSD 3.0: a comprehensive resource for the leaf senescence research community. Nucleic Acids Res. 48 D1069–D1075. 10.1093/nar/gkz898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Kim J. H., Kim J., Lyu J. I., Zhang Y., Guo H., et al. (2020b). ATM suppresses leaf senescence triggered by DNA double-strand break through epigenetic control of senescence-associated genes in Arabidopsis. New Phytol. 227 473–484. 10.1111/nph.16535 [DOI] [PubMed] [Google Scholar]
- Liang C., Wang Y., Zhu Y., Tang J., Hu B., Liu L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U S A 111 10013–10018. 10.1073/pnas.1321568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P. O., Kim H. J., Nam H. G. (2007). Leaf senescence. Annu. Rev. Plant. Biol. 58 115–136. 10.1146/annurev.arplant.57.032905.105316 [DOI] [PubMed] [Google Scholar]
- Lin Y. L., Sung S. C., Tsai H. L., Yu T. T., Radjacommare R., Usharani R., et al. (2011). The defective proteasome but not substrate recognition function is responsible for the null phenotypes of the Arabidopsis proteasome subunit RPN10. Plant Cell 23 2754–2773. 10.1105/tpc.111.086702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira B. S., Gramegna G., Trench B. A., Alves F. R. R., Silva E. M., Silva G. F. F., et al. (2017). Manipulation of a Senescence-Associated Gene Improves Fleshy Fruit Yield. Plant Physiol. 175 77–91. 10.1104/pp.17.00452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Zhang S., Zhou B., Luo X., Zhou X. F., Cai B., et al. (2019). The Histone H3K4 Demethylase JMJ16 Represses Leaf Senescence in Arabidopsis. Plant Cell 31 430–443. 10.1105/tpc.18.00693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wei W., Zhu W., Su L., Xiong Z., Zhou M., et al. (2017). Histone Deacetylase AtSRT1 Links Metabolic Flux and Stress Response in Arabidopsis. Mol. Plant 10 1510–1522. 10.1016/j.molp.2017.10.010 [DOI] [PubMed] [Google Scholar]
- Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389 251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Ma X., Zhang Y., Tureckova V., Xue G. P., Fernie A. R., Mueller-Roeber B., et al. (2018). The NAC Transcription Factor SlNAP2 Regulates Leaf Senescence and Fruit Yield in Tomato. Plant Physiol. 177 1286–1302. 10.1104/pp.18.00292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood K., El-Kereamy A., Kim S. H., Nambara E., Rothstein S. J. (2016). ANAC032 Positively Regulates Age-Dependent and Stress-Induced Senescence in Arabidopsis thaliana. Plant Cell Physiol. 57 2029–2046. 10.1093/pcp/pcw120 [DOI] [PubMed] [Google Scholar]
- Mao C., Lu S., Lv B., Zhang B., Shen J., He J., et al. (2017). A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis. Plant Physiol. 174 1747–1763. 10.1104/pp.17.00542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino-Ramirez L., Kann M. G., Shoemaker B. A., Landsman D. (2005). Histone structure and nucleosome stability. Exp. Rev. Proteomics 2 719–729. 10.1586/14789450.2.5.719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Clement G., Anne P., Routaboul J. M., Guiboileau A., Soulay F., et al. (2014). Stitching together the Multiple Dimensions of Autophagy Using Metabolomics and Transcriptomics Reveals Impacts on Metabolism, Development, and Plant Responses to the Environment in Arabidopsis. Plant Cell 26 1857–1877. 10.1105/tpc.114.124677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka D., Yasufuku T., Furuya T., Nanmori T. (2015). An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol. Biol. 87 565–575. 10.1007/s11103-015-0295-0 [DOI] [PubMed] [Google Scholar]
- Melo B. P., Fraga O. T., Silva J. C. F., Ferreira D. O., Brustolini O. J. B., Carpinetti P. A., et al. (2018). Revisiting the Soybean GmNAC Superfamily. Front. Plant Sci. 9:1864. 10.3389/fpls.2018.01864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Laun T., Zimmermann P., Zentgraf U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55 853–867. 10.1007/s11103-004-2142-6 [DOI] [PubMed] [Google Scholar]
- Miao Y., Zentgraf U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19 819–830. 10.1105/tpc.106.042705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. A. J., Shilatifard A. (2020). Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 52 1271–1281. 10.1038/s41588-020-00736-4 [DOI] [PubMed] [Google Scholar]
- Na J. K., Kim J. K., Kim D. Y., Assmann S. M. (2015). Expression of potato RNA-binding proteins StUBA2a/b and StUBA2c induces hypersensitive-like cell death and early leaf senescence in Arabidopsis. J. Exp. Bot. 66 4023–4033. 10.1093/jxb/erv207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahage I. S. P., Sakamoto S., Nagano M., Ishikawa T., Mitsuda N., Kawai-Yamada M., et al. (2020). An Arabidopsis NAC domain transcription factor, ATAF2, promotes age-dependent and dark-induced leaf senescence. Physiol. Plant 170 299–308. 10.1111/ppl.13156 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819 97–103. 10.1016/j.bbagrm.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Niu F., Cui X., Zhao P., Sun M., Yang B., Deyholos M. K., et al. (2020). WRKY42 transcription factor positively regulates leaf senescence through modulating SA and ROS synthesis in Arabidopsis thaliana. Plant J. 104 171–184. 10.1111/tpj.14914 [DOI] [PubMed] [Google Scholar]
- Oda-Yamamizo C., Mitsuda N., Sakamoto S., Ogawa D., Ohme-Takagi M., Ohmiya A. (2016). The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci. Rep. 6:23609. 10.1038/srep23609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak C., van Doorn W. G. (2005). Delay of Iris flower senescence by protease inhibitors. New Phytol. 165 473–480. 10.1111/j.1469-8137.2004.01226.x [DOI] [PubMed] [Google Scholar]
- Pandey R., Muller A., Napoli C. A., Selinger D. A., Pikaard C. S., Richards E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30 5036–5055. 10.1093/nar/gkf660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. S., Yao T., Seo J. S., Wong E. C. C., Mitsuda N., Huang C. H., et al. (2018). Arabidopsis NITROGEN LIMITATION ADAPTATION regulates ORE1 homeostasis during senescence induced by nitrogen deficiency. Nat. Plants 4 898–903. 10.1038/s41477-018-0269-8 [DOI] [PubMed] [Google Scholar]
- Park S. H., Jeong J. S., Seo J. S., Park B. S., Chua N. H. (2019). Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. New Phytol. 223 1447–1460. 10.1111/nph.15879 [DOI] [PubMed] [Google Scholar]
- Parkash J., Vaidya T., Kirti S., Dutt S. (2014). Translation initiation factor 5A in Picrorhiza is up-regulated during leaf senescence and in response to abscisic acid. Gene 542 1–7. 10.1016/j.gene.2014.03.032 [DOI] [PubMed] [Google Scholar]
- Phillips A. R., Suttangkakul A., Vierstra R. D. (2008). The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178 1339–1353. 10.1534/genetics.107.086199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao W., Sakuraba Y., Paek N. C. (2019). Transgenic expression of rice MYB102 (OsMYB102) delays leaf senescence and decreases abiotic stress tolerance in Arabidopsis thaliana. BMB Rep. 653–658. 10.5483/BMBRep.2019.52.11.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta M. R., Silva P. A., Mendes G. C., Alves J. R., Caetano H. D., Machado J. P., et al. (2016). The Stress-Induced Soybean NAC Transcription Factor GmNAC81 Plays a Positive Role in Developmentally Programmed Leaf Senescence. Plant Cell Physiol. 57 1098–1114. 10.1093/pcp/pcw059 [DOI] [PubMed] [Google Scholar]
- Poret M., Chandrasekar B., van der Hoorn R. A. L., Avice J. C. (2016). Characterization of senescence-associated protease activities involved in the efficient protein remobilization during leaf senescence of winter oilseed rape. Plant Sci. 246 139–153. 10.1016/j.plantsci.2016.02.011 [DOI] [PubMed] [Google Scholar]
- Qi T., Wang J., Huang H., Liu B., Gao H., Liu Y., et al. (2015). Regulation of Jasmonate-Induced Leaf Senescence by Antagonism between bHLH Subgroup IIIe and IIId Factors in Arabidopsis. Plant Cell 27 1634–1649. 10.1105/tpc.15.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines T., Shanks C., Cheng C. Y., McPherson D., Argueso C. T., Kim H. J., et al. (2016). The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J. 85 134–147. 10.1111/tpj.13097 [DOI] [PubMed] [Google Scholar]
- Ren T., Wang J., Zhao M., Gong X., Wang S., Wang G., et al. (2018). Involvement of NAC transcription factor SiNAC1 in a positive feedback loop via ABA biosynthesis and leaf senescence in foxtail millet. Planta 247 53–68. 10.1007/s00425-017-2770-0 [DOI] [PubMed] [Google Scholar]
- Reviron M. P., Vartanian N., Sallantin M., Huet J. C., Pernollet J. C., de Vienne D. (1992). Characterization of a Novel Protein Induced by Progressive or Rapid Drought and Salinity in Brassica napus Leaves. Plant Physiol. 100 1486–1493. 10.1104/pp.100.3.1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riester L., Koster-Hofmann S., Doll J., Berendzen K. W., Zentgraf U. (2019). Impact of Alternatively Polyadenylated Isoforms of ETHYLENE RESPONSE FACTOR4 with Activator and Repressor Function on Senescence in Arabidopsis thaliana L. Genes 10:2. 10.3390/genes10020091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Somssich I. E. (2001). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 28 123–133. 10.1046/j.1365-313x.2001.01131.x [DOI] [PubMed] [Google Scholar]
- Roussin-Leveillee C., Silva-Martins G., Moffett P. (2020). ARGONAUTE5 Represses Age-Dependent Induction of Flowering through Physical and Functional Interaction with miR156 in Arabidopsis. Plant Cell Physiol. 61 957–966. 10.1093/pcp/pcaa022 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Jeong J., Kang M. Y., Kim J., Paek N. C., Choi G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636. 10.1038/ncomms5636 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Kim D., Han S. H., Kim S. H., Piao W., Yanagisawa S., et al. (2020). Multilayered Regulation of Membrane-Bound ONAC054 Is Essential for Abscisic Acid-Induced Leaf Senescence in Rice. Plant Cell 32 630–649. 10.1105/tpc.19.00569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Piao W., Lim J. H., Han S. H., Kim Y. S., An G., et al. (2015). Rice ONAC106 Inhibits Leaf Senescence and Increases Salt Tolerance and Tiller Angle. Plant Cell Physiol. 56 2325–2339. 10.1093/pcp/pcv144 [DOI] [PubMed] [Google Scholar]
- Schelbert S., Aubry S., Burla B., Agne B., Kessler F., Krupinska K., et al. (2009). Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21 767–785. 10.1105/tpc.108.064089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M., Dimitrova M., Mancera-Martinez E., Geldreich A., Keller M., Ryabova L. A. (2013). TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 32 1087–1102. 10.1038/emboj.2013.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chetelat A., Cubas P., Farmer E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi-Schwager M., Lers A., Samach A., Guy C. L., Porat R. (2010). Overexpression of the CBF2 transcriptional activator in Arabidopsis delays leaf senescence and extends plant longevity. J. Exp. Bot. 61 261–273. 10.1093/jxb/erp300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. (2005). The discovery of split genes and RNA splicing. Trends Biochem. Sci. 30 279–281. 10.1016/j.tibs.2005.04.002 [DOI] [PubMed] [Google Scholar]
- Shen Y., Lei T., Cui X., Liu X., Zhou S., Zheng Y., et al. (2019). Arabidopsis histone deacetylase HDA15 directly represses plant response to elevated ambient temperature. Plant J. 100 991–1006. 10.1111/tpj.14492 [DOI] [PubMed] [Google Scholar]
- Shi H., Reiter R. J., Tan D. X., Chan Z. (2015). INDOLE-3-ACETIC ACID INDUCIBLE 17 positively modulates natural leaf senescence through melatonin-mediated pathway in Arabidopsis. J. Pineal. Res. 58 26–33. 10.1111/jpi.12188 [DOI] [PubMed] [Google Scholar]
- Shin D., Lee S., Kim T. H., Lee J. H., Park J., Lee J., et al. (2020). Natural variations at the Stay-Green gene promoter control lifespan and yield in rice cultivars. Nat. Commun. 11:2819. 10.1038/s41467-020-16573-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S. H., Bleecker A. B. (2001). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 2001:re22. 10.1126/stke.2001.113.re22 [DOI] [PubMed] [Google Scholar]
- Shiu S. H., Karlowski W. M., Pan R., Tzeng Y. H., Mayer K. F., Li W. H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16 1220–1234. 10.1105/tpc.020834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K., Yang W. (2017). E3 Ubiquitin Ligases: Ubiquitous Actors in Plant Development and Abiotic Stress Responses. Plant Cell Physiol. 58 1461–1476. 10.1093/pcp/pcx071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Huang H., Wang J., Liu B., Qi T., Xie D. (2017). MYC5 is Involved in Jasmonate-Regulated Plant Growth, Leaf Senescence and Defense Responses. Plant Cell Physiol. 58 1752–1763. 10.1093/pcp/pcx112 [DOI] [PubMed] [Google Scholar]
- Song Y., Jiang Y., Kuai B., Li L. (2018). CIRCADIAN CLOCK-ASSOCIATED 1 Inhibits Leaf Senescence in Arabidopsis. Front. Plant Sci. 9:280. 10.3389/fpls.2018.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Yang C., Gao S., Zhang W., Li L., Kuai B. (2014). Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 7 1776–1787. 10.1093/mp/ssu109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Ji X., Jia Y., Huo D., Si S., Zeng L., et al. (2020). LreEF1A4, a Translation Elongation Factor from Lilium regale, Is Pivotal for Cucumber Mosaic Virus and Tobacco Rattle Virus Infections and Tolerance to Salt and Drought. Int. J. Mol. Sci. 21:6. 10.3390/ijms21062083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Makino A. (2013). Translational downregulation of RBCL is operative in the coordinated expression of Rubisco genes in senescent leaves in rice. J. Exp. Bot. 64 1145–1152. 10.1093/jxb/ers398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak H., Negi S., Gupta A., Ganapathi T. R. (2018). A stress associated NAC transcription factor MpSNAC67 from banana (Musa x paradisiaca) is involved in regulation of chlorophyll catabolic pathway. Plant Physiol. Biochem. 132 61–71. 10.1016/j.plaphy.2018.08.020 [DOI] [PubMed] [Google Scholar]
- Takasaki H., Maruyama K., Takahashi F., Fujita M., Yoshida T., Nakashima K., et al. (2015). SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 84 1114–1123. 10.1111/tpj.13067 [DOI] [PubMed] [Google Scholar]
- Tang D., Innes R. W. (2002). Overexpression of a kinase-deficient form of the EDR1 gene enhances powdery mildew resistance and ethylene-induced senescence in Arabidopsis. Plant J. 32 975–983. 10.1046/j.1365-313x.2002.01482.x [DOI] [PubMed] [Google Scholar]
- Tian F., Yu J., Zhang Y., Xie Y., Wu B., Miao Y. (2019). MORF9 Functions in Plastid RNA Editing with Tissue Specificity. Int. J. Mol. Sci. 20:18. 10.3390/ijms20184635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T., Kato W., Asada Y., Sako K., Sato T., Sonoda Y., et al. (2009). DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J. Plant Res. 122 633–643. 10.1007/s10265-009-0252-6 [DOI] [PubMed] [Google Scholar]
- Uauy C., Distelfeld A., Fahima T., Blechl A., Dubcovsky J. (2006). A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314 1298–1301. 10.1126/science.1133649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M., Matsui A., Nakamura T., Abe T., Sunaoshi Y., Shimada H., et al. (2018). Versatility of HDA19-deficiency in increasing the tolerance of Arabidopsis to different environmental stresses. Plant Signal Behav. 13:e1475808. 10.1080/15592324.2018.1475808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uji Y., Akimitsu K., Gomi K. (2017). Identification of OsMYC2-regulated senescence-associated genes in rice. Planta 245 1241–1246. 10.1007/s00425-017-2697-5 [DOI] [PubMed] [Google Scholar]
- Upadhyay R. K., Soni D. K., Singh R., Dwivedi U. N., Pathre U. V., Nath P., et al. (2013). SlERF36, an EAR-motif-containing ERF gene from tomato, alters stomatal density and modulates photosynthesis and growth. J. Exp. Bot. 64 3237–3247. 10.1093/jxb/ert162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Arroyo B., Diaz-Mendoza M., Gandullo J., Gonzalez-Melendi P., Santamaria M. E., Dominguez-Figueroa J. D., et al. (2016). HvPap-1 C1A protease actively participates in barley proteolysis mediated by abiotic stresses. J. Exp. Bot. 67 4297–4310. 10.1093/jxb/erw212 [DOI] [PubMed] [Google Scholar]
- Wang H., Schippers J. H. M. (2019). The Role and Regulation of Autophagy and the Proteasome During Aging and Senescence in Plants. Genes 10:4. 10.3390/genes10040267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. L., Zhang Y., Wang T., Yang Q., Yang Y., Li Z., et al. (2021). An Alternative Splicing Variant of PtRD26 Delays Leaf Senescence by Regulating Multiple NAC Transcription Factors in Populus. Plant Cell 2021:46. 10.1093/plcell/koab046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. W. (2014). Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 65 4723–4730. 10.1093/jxb/eru246 [DOI] [PubMed] [Google Scholar]
- Wang S., Lei C., Wang J., Ma J., Tang S., Wang C., et al. (2017). SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J. Exp. Bot. 68 899–913. 10.1093/jxb/erx001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. H., Lim J. H., Kim S. S., Cho S. H., Yoo S. C., Koh H. J., et al. (2015). Mutation of SPOTTED LEAF3 (SPL3) impairs abscisic acid-responsive signalling and delays leaf senescence in rice. J. Exp. Bot. 66 7045–7059. 10.1093/jxb/erv401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. W., Lu L., Wang D., Thompson J. E. (2001). Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eucaryotic translation initiation factor 5A from tomato. J. Biol. Chem. 276 17541–17549. 10.1074/jbc.M008544200 [DOI] [PubMed] [Google Scholar]
- Wang X., Gao J., Gao S., Song Y., Yang Z., Kuai B. (2019). The H3K27me3 demethylase REF6 promotes leaf senescence through directly activating major senescence regulatory and functional genes in Arabidopsis. PLoS Genet. 15:e1008068. 10.1371/journal.pgen.1008068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cui X., Yang B., Xu S., Wei X., Zhao P., et al. (2020). WRKY55 transcription factor positively regulates leaf senescence and the defense response by modulating the transcription of genes implicated in the biosynthesis of reactive oxygen species and salicylic acid in Arabidopsis. Development 147:16. 10.1242/dev.189647 [DOI] [PubMed] [Google Scholar]
- Wen Z., Mei Y., Zhou J., Cui Y., Wang D., Wang N. N. (2020). SAUR49 Can Positively Regulate Leaf Senescence by Suppressing SSPP in Arabidopsis. Plant Cell Physiol. 61 644–658. 10.1093/pcp/pcz231 [DOI] [PubMed] [Google Scholar]
- Woo H. R., Goh C. H., Park J. H., Teyssendier de la Serve B., Kim J. H., Park Y., I, et al. (2002). Extended leaf longevity in the ore4-1 mutant of Arabidopsis with a reduced expression of a plastid ribosomal protein gene. Plant J. 31 331–340. 10.1046/j.1365-313x.2002.01355.x [DOI] [PubMed] [Google Scholar]
- Woo H. R., Kim H. J., Lim P. O., Nam H. G. (2019). Leaf Senescence: Systems and Dynamics Aspects. Annu. Rev. Plant Biol. 70 347–376. 10.1146/annurev-arplant-050718-095859 [DOI] [PubMed] [Google Scholar]
- Woo H. R., Kim H. J., Nam H. G., Lim P. O. (2013). Plant leaf senescence and death - regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 126(Pt 21) 4823–4833. 10.1242/jcs.109116 [DOI] [PubMed] [Google Scholar]
- Woo H. R., Kim J. H., Kim J., Kim J., Lee U., Song I. J., et al. (2010). The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot. 61 3947–3957. 10.1093/jxb/erq206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H. R., Koo H. J., Kim J., Jeong H., Yang J. O., Lee I. H., et al. (2016). Programming of Plant Leaf Senescence with Temporal and Inter-Organellar Coordination of Transcriptome in Arabidopsis. Plant Physiol. 171 452–467. 10.1104/pp.15.01929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Allu A. D., Garapati P., Siddiqui H., Dortay H., Zanor M. I., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24 482–506. 10.1105/tpc.111.090894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Li B., Iwakawa H. O., Pan Y., Tang X., Ling-Hu Q., et al. (2020). Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature 581 89–93. 10.1038/s41586-020-2231-y [DOI] [PubMed] [Google Scholar]
- Wu X., Ding D., Shi C., Xue Y., Zhang Z., Tang G., et al. (2016). microRNA-dependent gene regulatory networks in maize leaf senescence. BMC Plant Biol. 16:73. 10.1186/s12870-016-0755-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D., Cui Y., Xu F., Xu X., Gao G., Wang Y., et al. (2015). SENESCENCE-SUPPRESSED PROTEIN PHOSPHATASE Directly Interacts with the Cytoplasmic Domain of SENESCENCE-ASSOCIATED RECEPTOR-LIKE KINASE and Negatively Regulates Leaf Senescence in Arabidopsis. Plant Physiol. 169 1275–1291. 10.1104/pp.15.01112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Hu Z., Zhu Z., Dong T., Zhao Z., Cui B., et al. (2014). Overexpression of a novel MADS-box gene SlFYFL delays senescence, fruit ripening and abscission in tomato. Sci. Rep. 4:4367. 10.1038/srep04367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Huhn K., Brandt R., Potschin M., Bieker S., Straub D., et al. (2014). REVOLUTA and WRKY53 connect early and late leaf development in Arabidopsis. Development 141 4772–4783. 10.1242/dev.117689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A. L., Bassham D. C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42 535–546. 10.1111/j.1365-313X.2005.02397.x [DOI] [PubMed] [Google Scholar]
- Xu F., Meng T., Li P., Yu Y., Cui Y., Wang Y., et al. (2011). A soybean dual-specificity kinase, GmSARK, and its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol. 157 2131–2153. 10.1104/pp.111.182899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Wang X., Chen J. (2010). Overexpression of the Rap2.4f transcriptional factor in Arabidopsis promotes leaf senescence. Sci. China Life Sci. 53 1221–1226. 10.1007/s11427-010-4068-3 [DOI] [PubMed] [Google Scholar]
- Xu T., Kim B. M., Kwak K. J., Jung H. J., Kang H. (2016). The Arabidopsis homolog of human minor spliceosomal protein U11-48K plays a crucial role in U12 intron splicing and plant development. J. Exp. Bot. 67 3397–3406. 10.1093/jxb/erw158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Umehara M. (2015). Possible Roles of Strigolactones during Leaf Senescence. Plants 4 664–677. 10.3390/plants4030664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Chen Q., Cui X., Zhao P., Gao S., Yang B., et al. (2021). Ectopic overexpression of a membrane-tethered transcription factor gene NAC60 from oilseed rape positively modulates programmed cell death and age-triggered leaf senescence. Plant J. 105 600–618. 10.1111/tpj.15057 [DOI] [PubMed] [Google Scholar]
- Yan J., Tong T., Li X., Chen Q., Dai M., Niu F., et al. (2018). A Novel NAC-Type Transcription Factor, NAC87, from Oilseed Rape Modulates Reactive Oxygen Species Accumulation and Cell Death. Plant Cell Physiol. 59 290–303. 10.1093/pcp/pcx184 [DOI] [PubMed] [Google Scholar]
- Yan Z., Jia J., Yan X., Shi H., Han Y. (2017). Arabidopsis KHZ1 and KHZ2, two novel non-tandem CCCH zinc-finger and K-homolog domain proteins, have redundant roles in the regulation of flowering and senescence. Plant Mol. Biol. 95 549–565. 10.1007/s11103-017-0667-8 [DOI] [PubMed] [Google Scholar]
- Yang L., Ye C., Zhao Y., Cheng X., Wang Y., Jiang Y. Q., et al. (2018). An oilseed rape WRKY-type transcription factor regulates ROS accumulation and leaf senescence in Nicotiana benthamiana and Arabidopsis through modulating transcription of RbohD and RbohF. Planta 247 1323–1338. 10.1007/s00425-018-2868-z [DOI] [PubMed] [Google Scholar]
- Yang S. D., Seo P. J., Yoon H. K., Park C. M. (2011). The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23 2155–2168. 10.1105/tpc.111.084913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Wang X., Ji L., Yi Z., Fu C., Ran J., et al. (2015). Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep. 34 943–958. 10.1007/s00299-015-1756-2 [DOI] [PubMed] [Google Scholar]
- Yang Z., Wang C., Qiu K., Chen H., Li Z., Li X., et al. (2020). The transcription factor ZmNAC126 accelerates leaf senescence downstream of the ethylene signalling pathway in maize. Plant Cell Environ. 43 2287–2300. 10.1111/pce.13803 [DOI] [PubMed] [Google Scholar]
- Yoon H. K., Kim S. G., Kim S. Y., Park C. M. (2008). Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol. Cells 25 438–445. [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16 2967–2983. 10.1105/tpc.104.025395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zhang Y., Di C., Zhang Q., Zhang K., Wang C., et al. (2016). JAZ7 negatively regulates dark-induced leaf senescence in Arabidopsis. J. Exp. Bot. 67 751–762. 10.1093/jxb/erv487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yujun R., Wanzhen W., Dirk S., Daguang C., Ying M. (2019). MicroRNA840 positively regulates plant senescence in Arabidopsis by interfering PPR and WHIRLY3 expression at posttranscriptional and translational levels. Chin. Soc. Plant Biol. 2019:1. [Google Scholar]
- Zentgraf U., Laun T., Miao Y. (2010). The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 89 133–137. 10.1016/j.ejcb.2009.10.014 [DOI] [PubMed] [Google Scholar]
- Zhan X., Qian B., Cao F., Wu W., Yang L., Guan Q., et al. (2015). An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 6:8139. 10.1038/ncomms9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Zhu Z., Gao J., Zhou X., Zhu S., Wang X., et al. (2021). The NPR1-WRKY46-WRKY6 signaling cascade mediates probenazole/salicylic acid-elicited leaf senescence in Arabidopsis thaliana. J. Integr. Plant Biol. 63 924–936. 10.1111/jipb.13044 [DOI] [PubMed] [Google Scholar]
- Zhang J., Gao J., Zhu Z., Song Y., Wang X., Wang X., et al. (2020). MKK4/MKK5-MPK1/MPK2 cascade mediates SA-activated leaf senescence via phosphorylation of NPR1 in Arabidopsis. Plant Mol. Biol. 102 463–475. 10.1007/s11103-019-00958-z [DOI] [PubMed] [Google Scholar]
- Zhang K., Halitschke R., Yin C., Liu C. J., Gan S. S. (2013). Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. U S A 110 14807–14812. 10.1073/pnas.1302702110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Wang W. Q., Zhang G. L., Kaminek M., Dobrev P., Xu J., et al. (2010). Senescence-inducible expression of isopentenyl transferase extends leaf life, increases drought stress resistance and alters cytokinin metabolism in cassava. J. Integr. Plant Biol. 52 653–669. 10.1111/j.1744-7909.2010.00956.x [DOI] [PubMed] [Google Scholar]
- Zhang X., Ju H. W., Chung M. S., Huang P., Ahn S. J., Kim C. S. (2011). The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol. 52 138–148. 10.1093/pcp/pcq180 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Gao Y., Wang H. L., Kan C., Li Z., Yang X., et al. (2021). Verticillium dahliae secretory effector PevD1 induces leaf senescence by promoting ORE1-mediated ethylene biosynthesis. Mol. Plant 2021:14. 10.1016/j.molp.2021.07.014 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu J., Chai J., Xing D. (2016). Mitogen-activated protein kinase 6 mediates nuclear translocation of ORE3 to promote ORE9 gene expression in methyl jasmonate-induced leaf senescence. J. Exp. Bot. 67 83–94. 10.1093/jxb/erv438 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yin S., Tu Y., Mei H., Yang Y. (2020). A novel microRNA, SlymiR208, promotes leaf senescence via regulating cytokinin biosynthesis in tomato. Physiol. Plant 169 143–155. 10.1111/ppl.13068 [DOI] [PubMed] [Google Scholar]
- Zhao D., Derkx A. P., Liu D. C., Buchner P., Hawkesford M. J. (2015). Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 17 904–913. 10.1111/plb.12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Ma J., Li L., Fan S., Guo Y., Song M., et al. (2016). GhNAC12, a neutral candidate gene, leads to early aging in cotton (Gossypium hirsutum L). Gene 576(1 Pt 2), 268–274. 10.1016/j.gene.2015.10.042 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Shilatifard A. (2019). Epigenetic modifications of histones in cancer. Genome Biol. 20:245. 10.1186/s13059-019-1870-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Ding Y., Sun X., Xie S., Wang D., Liu X., et al. (2016). Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 67 1703–1713. 10.1093/jxb/erv562 [DOI] [PubMed] [Google Scholar]
- Zhou C., Cai Z., Guo Y., Gan S. (2009). An arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 150 167–177. 10.1104/pp.108.133439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Zhao J., Cai J., Patil S. B. (2017). UBIQUITIN-SPECIFIC PROTEASES function in plant development and stress responses. Plant Mol. Biol. 94 565–576. 10.1007/s11103-017-0633-5 [DOI] [PubMed] [Google Scholar]
- Zhou J., Lu D., Xu G., Finlayson S. A., He P., Shan L. (2015). The dominant negative ARM domain uncovers multiple functions of PUB13 in Arabidopsis immunity, flowering, and senescence. J. Exp. Bot. 66 3353–3366. 10.1093/jxb/erv148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Jiang Y., Yu D. (2011). WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 31 303–313. 10.1007/s10059-011-0047-1 [DOI] [PMC free article] [PubMed] [Google Scholar]


