Abstract
Background and objectives
Remarkable infectivity of severe acute respiratory syndrome-coronavirus 2 (SARS-CoV2) is due to the rapid emergence of various strains which enable the virus to ruling the world. Over the course of SARS-CoV2 pandemic, the scientific communities worldwide are responding to newly emerging genetic variants. However, mechanism behind the persistent infection of these variants is still not known due to the paucity of study of these variants at molecular level. In this scenario, computational methods have immense utility in understanding the molecular and functional properties of different variants.
Methods
The various mutants (MTs) of SpikeS1 receptor binding domain (RBD) of highly infectious SARS-CoV2 strains were manifested and elucidated the protein structure and binding strength using molecular dynamics (MD) simulation and protein-protein docking approaches.
Results
MD simulation study showed that all MTs exhibited stable structures with altered functional properties. Furthermore, the binding strength of different MTs along with WT (wildtype) was revealed that MTs showed differential binding affinities to host protein with high binding strength exhibited by V367F and V483A MTs.
Conclusion
Hence, this study shed light on the molecular basis of infection caused by different variants of SARS-CoV2, which might play an important role in to cease the transmission and pathogenesis of virus and also implicate in rational designing of a specific drug.
Keywords: SARS-CoV2, Mutant strains, Molecular dynamics, Network analysis, Protein-protein interaction
List of abbreviations
- ACE2
angiotensin converting enzyme2
- COVID-19
coronavirus disease-2019
- DSSP
dictionary of secondary structure of protein
- ED
essential dynamics
- GROMACS
groningen machine for chemical simulation
- HADDOCK
high ambiguity driven docking
- MDS
molecular dynamics simulation
- MT
mutant
- PCA
principal component analysis
- PDB
protein data bank
- ProSA
protein structure analysis
- QMEAN
qualitative model energy analysis
- RBD
receptor binding domain
- Rg
radius of gyration
- RIN
residues interaction network
- RMSD
root mean square deviation
- RMSF
root mean square fluctuation
- SARS-CoV2
severe acute respiratory syndrome-coronavirus 2
- SASA
solvent accessible surface area
- SAVES
structure analysis and verification server
- WT
wildtype
1. Introduction
Near the end of 2019, new public health crises arise from Wuhan city of China due to the outbreak of nCoV (novel coronavirus) or SARS-CoV2, which spread worldwide and named COVID-19 (coronavirus disease 2019) [1]. Till now (September 2021), the cumulative number of total confirmed cases and deaths of COVID-19 reached up to ∼228 and ∼4.6 million (https://www.worldometers.info/coronavirus/), respectively (Fig. S1A and B; Tables S1 and S2). During the first wave of SARS-CoV2, most of the confirmed cases are observed in the USA (∼2.64 millions) (Fig. S1C, Table S1), while during the second wave, its number reached ∼20.06 million (Fig. S1E) [2]. To date, ∼43.35 million confirmed cases were observed in the USA alone, which are the highest number of cases arisen in any country in entire world [2]. However, the cumulative confirmed COVID-19 deaths during first (∼575.02 per million deaths) and second waves (∼1226.54 deaths per million) of infections were reported highest in Italy (Fig. S1D and F) [3]. While in Brazil, ∼2778.63 deaths per million were observed, which are the highest number of deaths per million estimated by any country throughout the world (Table S2). Rapid global spread and transmission of COVID-19 provide the virus with substantial opportunities for the natural selection of favorable mutations [4]. Mutation variants in SARS-CoV2 genome have raised serious concern about changes in infectivity [5]. The emergence of new strains driven by the adaptive mutation indicate its instability over a period of time and can contribute toward its virulence. This rapid evolution seems to be a common problem that shows the gravity of coronavirus infection as a communicable disease and attract the attention of the scientific community as a matter of serious concern worldwide [6]. Mostly, different strains are arising due to mutations in the receptor binding domain (RBD) of SpikeS1 protein [7]. SpikesS1 protein of virus interacts with host angiotensin converting enzyme2 (ACE2) that enable the virus to enter into the host cell [8]. V367F (Valine 367 to Phenylalanine) mutation in the RBD of SpikeS1 protein was reported from the strains found in Wuhan, Shenzhen, Hong Kong and France [9,10]. R408I (Arginine 408 to Isoleucine) mutant isolated from India [11]. In addition, mutations such as G476S (Glycine 476 to Serine) and V483A (Valine 483 to Alanine) were reported in the United States [12]. Another strain in which Asparagine positioned at 501 of RBD is substituted by Tyrosine (N501Y) arose in United Kingdom and South Africa [13,14]. These mutations (V367F, R408I, G476S, V483A and N501Y) were mainly located in the SpikeS1 RBD domain. Of these five, three mutations such as G476S, V483A and N501Y occur at the binding interface of RBD (C-terminal side) that interact with PD (peptidase domain) of the host ACE2 receptor [15]. The rest of mutations V367F and R408I, mostly affected the overall topology and stability of RBD as these mutations present at the loop regions of N-terminal and turn that connect β-sheet3 and 4, respectively [16]. These mutations (V367F, R408I, G476S, V483A and N501Y) indirectly assist in the stable binding of RBD to the host ACE2 receptor [15]. In addition to the above mutations, there are various other mutations found in the SpikeS1 protein beyond the RBD domain that also contributed to the virus infectivity [16]. Since targeting the mutations in RBD domain provide the first step to halt the virus infection, therefore the current study is mainly focused on the RBD mutations. To unravel the stability and dynamics of highly infectious strains or RBD mutants (MTs), long run molecular dynamics (MD) simulations were performed followed by elucidating the key residues involved in signalling through residues interaction network (RIN) approach. Furthermore, the infectiveness of above MTs were assessed through protein-protein (ACE2-RBD) docking of both WT and MT RBD proteins. We observed that all MTs showed similar dynamic but differential structural properties. Moreover, RBD MTs such as V367F and V483A showed high binding affinities to the host ACE2 receptor protein. Hence, the work in the present study will help in the creation of a more specific inhibitor or drug by taking the above residues in consideration.
2. Materials and methods
2.1. 3D structures preparation and validation
The tertiary structure of RBD of Spike S1 protein was taken from RCSB-PDB (Research collaboration for structural bioinformatics-protein data bank), which was experimentally resolved in complexed form (PDB id: 6M17) [16,17]. The structure was prepared in PyMOL (The PyMOL Molecular Graphics System, Version 1.3 Schrodinger, LLC) and all other structures except RBD were excluded. Distinct missense mutations which represented different strains of SARS-CoV2 such as V367F, R408I, G476S, V483A, N501Y were prepared in PyMOL as previously described [18]. 3D structures of all mutants (V367F, R408I, G476S, V483A and N501Y) along with wildtype were energy minimized using GROMACS [19]. Energy minimized protein models were subjected to validation by different quality estimation tools. Stereochemical properties, as well as local and global qualities of all models were monitored through PROCHECK module of SAVES (Structure analysis and verification server), ProSA (Protein structure analysis; https://prosa.services.came.sbg.ac.at/prosa.php) and QMEAN (Qualitative model energy analysis; https://swissmodel.expasy.org/qmean/) web servers, respectively [20], [21], [22].
2.2. Molecular dynamics simulation
Molecular dynamics (MD) simulations of 150 ns for each WT and MTs were conducted using GROMACS in conjunction with Amber force field [19,23]. Initially, GROMACS readable coordinates and topology files were generated through “pdb2gmx” module and all systems (WT and MTs) were solvated using “editconf” and solvate scripts. Systems solvation were accomplished by immersing all WT and MT proteins into triclinic box with TIP3P water model. After that, all systems were neutralized by adding sodium and chloride counterions using “genion” module followed by energy minimization through the steepest-descent method. Hence after, two equilibration steps as NVT (constant number of particles, volume, and temperature) and NPT (constant number of particles, pressure, and temperature) ensembles were performed for 100 and 500 ps at a temperature of 300 K and pressure of 1bar, respectively. The temperature and pressure of all the systems were carried by v‐rescale (a modified Berendsen method) and Parrinello‐Rahman methods, respectively. Long range electrostatic interactions were preserved by using Particle Mesh Ewald (PME) methods and all bonds were constraint using Linear constraint solver (LINCS) algorithm. Finally, the production run of 150 ns was performed in which time steps of 2fs were applied. Resultant MD trajectories were analyzed by using gmx energy, gmx rms, gmx rmsf, gmx gyrate, gmx sasa and gmx hbond modules of GROMACS. Secondary structures analysis was done through the dictionary of secondary structure of protein (DSSP) approach using do_dssp script [24].
2.3. Essential dynamics
Essential dynamics (ED) is an important parameter, used to generate coordinates of subspace in which the motion of a protein is likely to take place that determines its biological function [25]. A series of eigenvectors with respective eigenvalues were generated after the construction and diagonalization of the covariance matrix of backbone atoms from all WT and MT systems. We restricted our interpretation with backbone atoms in order to avoid statistical noise and utilized stabled trajectories of last 25 ns of all WT and MT systems for our ED analysis. Simultaneously, to avoid the biologically misinterpretation of PCs we calculated cosine values of first three PCs that should be ≤0.2 [26]. First three principal components (PC1, PC2 and PC3) or eigenvectors were used which usually accompanied with maximum motions that are biologically significant [27]. ED analysis was performed by gmx covar, gmx analyse and gmx anaeig modules of GROMACS.
2.4. Residues network analysis
Residues interacting networks (RIN) of WT and MTs were predicted on MD optimized protein structure using NAPS (Network Analysis of Protein Structure) webserver [28]. The server facilitated qualitative and quantitative analyses of RIN and protein contact network was constructed based on physiochemical properties of residues in which different centralities were measured. We restrict our analysis with Cα atoms where the residue is treated as node with an unweighted edge drawn within distance of 7 Å connecting other residues in the given network. NAPS administered different centrality measures, such as degree (CD), closeness (CC), and betweenness (CB). CD is defined as the number of node link with the central node as mentioned in Eq. (1), while CC is the average farness from the central node to other nodes within the network as mentioned in Eqs. (1) and (2). CB is depending upon the shortest path between the nodes. It is the ratio of shortest routes from one node to other passes through u and the entire shortest path within the network as described in Eq. (3).
| (1) |
| (2) |
| (3) |
Whereas, A = adjacency matrix; V= set of all nodes; N= number of nodes in the network; dist = shortest path distance between nodes u and = shortest path between s and t; σst(u) = shortest path between s and t passing through u.
2.5. Protein-protein docking
Protein-protein (p-p) docking of ACE2 and all WT and MTs SpikeS1 RBD were executed through HADDOCK (High Ambiguity Driven protein-protein DOCKing) webserver [29]. Active residues which were identified through available experimental complex structures from PDB, used as constraint to generate AIRs (Ambiguous Interaction Restraints) during docking [16]. We have used the MD refined structures of ACE2 and RBD MTs for protein-protein docking analysis. HADDOCK carried p-p docking in 3 successive stages as previously described [30]. Briefly, energy minimization steps were performed initially, followed by simulated annealing refinement and finally MD refinement with solvent. P-p complex structures obtained from the final stage were clustered and HADDOCK scores were calculated as given in Eqs. (4)–(6).
| (4) |
| (5) |
| (6) |
In the above equations, Evdw, Eelec, EAIR, Edesolv, Edata, BSA, represented van der Waals, electrostatic, AIR, desolvation energies and energy of other restraint data and buried surface area, respectively. Best docked complexes with the lowest HADDOCK scores from all ACE-WT and -MT complexes were finally analyzed. Further, p-p docking of ACE2 and all WT and MTs SpikeS1 RBD were also predicted through ClusPro and pyDOCKWEB servers [31,32]. Protein-protein interaction visualizations at 3D and 2D levels and H-bonding were analyzed in PyMOL (The PyMOL Molecular Graphics System, Version 1.3 Schrodinger, LLC) and DIMPLOT module of LigPlot+ v2.2.4, respectively [33]. Furthermore, the binding strength of various ACE2-WT and -MTs complexes were also verified by using mCSM-PPI2 server [34]. mCSM-PPI2 is novel computational tool that accurately predicts the impacts of missense mutations in p-p binding affinity. It used graph-based structural signatures (mCSM) to represent the wild-type residue environment, inter-residue non-covalent interaction networks, and evolutionary information, complex network metrics, and energetic terms to generate an optimized predictor.
3. Results
3.1. WT and MTs demonstrated similar dynamic behaviors
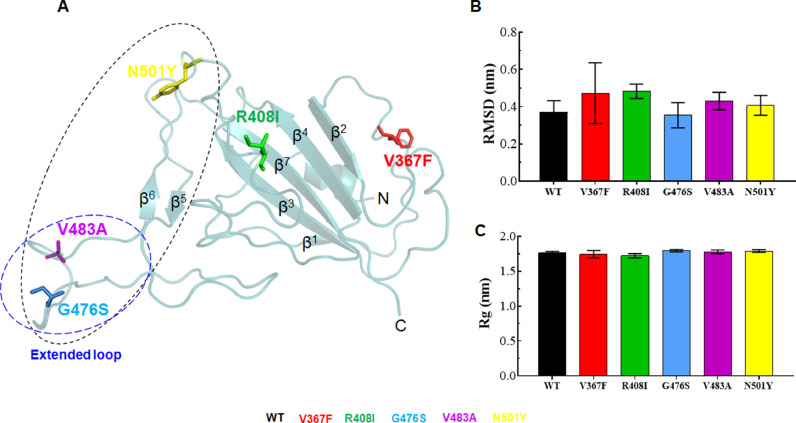
The tertiary structure of RBD comprized about 181 amino acids long polypeptide and have seven β-sheets (Fig. 1 A). An extended loop was present near C-terminal that facilitated its (RBD) interaction with protein partner (ACE2). Various MT models were generated in PyMOL by replacing Valine367 with Phenylalanine (V367F), Arginine408 with Isoleucine (R408I), Glutamine476 with Serine (G476S), Valine483 with Alanine (V483A) and Asparagine501 with Tyrosine (N501Y) in WT 3D structure. WT and MT model structures were energy minimized and found that all models were exhibited the lowest values of minimum energies (data not shown). V367F and R408I MTs were present in between β-sheets 1 and 2, β-sheet 3 and 4, respectively (Fig. 1A). Additionally, an extended loop was present near to C-terminal harboured 2 mutations such as G476S and V483A, while N501Y mutation was located near to the end of C-terminal. The extended loop and the region near C-terminal of SpikeS1 RBD interacts with the peptidase domain (PD) of ACE receptor [15]. The structure validations of WT and newly constructed MT models were inspected by measuring the Phi/Psi angles in the Ramachandran plot and found that all 3D structures exhibited 68.5%, 29.5% and 2% of residues to be placed in favorable, allowed and disallowed regions, respectively (Table S3). Moreover, ProSA and QMEAN servers were also executed to analyse the local and global geometry of WT and MT structures and found that all WT and MT proteins exhibited ProSA and QMEAN values ranging from -5.15 to -5.25 and -7.33 to -8.15, respectively (Table S3). The negligible number of residues in disallowed regions and lowest values of ProSA and QMEAN scores indicated that all WT and MT protein structures had better stereochemical and geometrical properties.
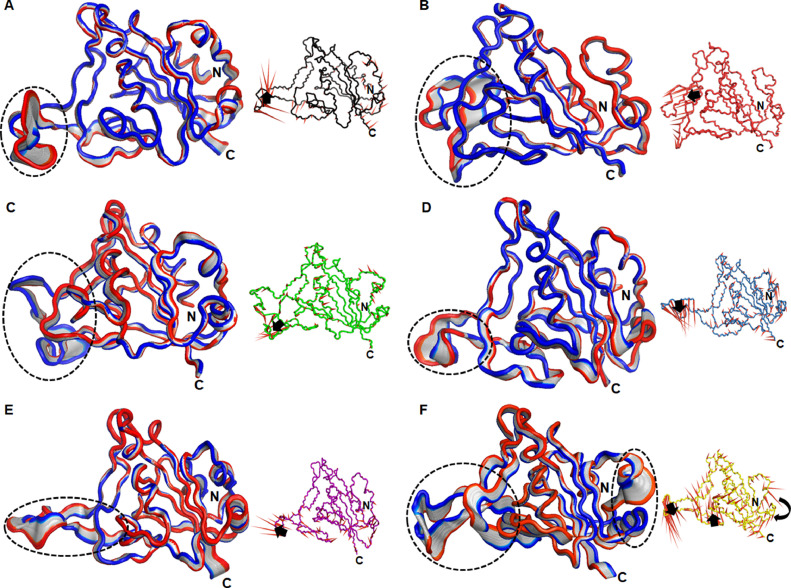
Fig. 1.
Tertiary structure, RMSD and Rg analyses. (A) Tertiary structure of SpikeS1 RBD of SARS-CoV2, (B) Root mean square deviation at function of time and (C) Radius of gyration at function of time in nanoseconds. 3D structure of RBD was shown in cartoon mode with cyan color and different mutations V367F, R408I, G476S, V483A and N501Y were depicted in stick mode. Region interacting with host ACE2 of SpikeS1 RBD and extended loop were highlighted in back and blue dotted circles, respectively. Different sheets were labelled as β1-β7. WT, V367F, R408I, G476S, V483A and N501Y MTs were labelled in black, red, green, blue, magenta and yellow color, respectively.
Preliminary analyses of effects of different mutations on 3D structure were predicted through MUpro [35] and I-mutant [36] servers and found that all mutant except N501Y showed decreased structure stabilities (Tables S4 and S5). Further, detailed structural stability and dynamics were examined through MD simulations. To assess the structural stabilities of WT and MTs (V367F, R408I, G476S, V483A and N501Y), root mean square deviation (RMSD) of protein backbone with respect to equilibrium structures were calculated and found that average RMSD values of ∼0.37 for WT, ∼0.47 for V367F, ∼0.48 for R408I, ∼0.35 for G476S, ∼0.43 for V483A and ∼0.40 nm for N501Y MTs were existed (Fig. 1B). Further, RMSDs of all WT and MTs were equilibrated after 100 ns time and showed similar pattern (Fig. S2A). Moreover, V367F and R408I MTs displayed higher deviations, whereas G476S MT exhibited lower deviation then WT, V483A and N501Y MTs (Fig. 1B). RMSD of all MTs and WT showed a similar and consistent pattern. Protein compactness or globularity was measured by employing the radius of gyration (Rg) and found that average Rg values of ∼1.76, ∼1.74, ∼1.72, ∼1.79, ∼1.77 and ∼1.78 nm were obtained for WT, V367F, R408I, G476S, V483A and N501Y MTs, respectively (Fig. 1C). Rgs were slightly reduced in V367F and R408I MTs, whereas G476S, V483A and N501Y MTs showed slightly higher Rg then WT. Further, the Rg patterns of all MTs and WT were stabilized after ∼125 ns time and showed similar behavior (Fig. S2B). Taken together, all MTs and WT showed similar pattern of RMSD and Rg, indicating that both WT and MT proteins exhibited similar dynamic behaviors.
3.2. Root mean square fluctuations analyses of WT and MTs
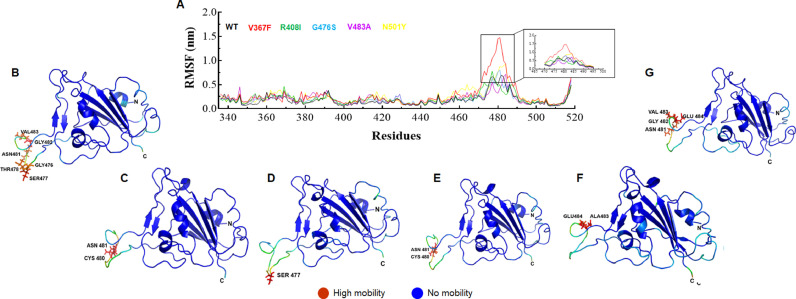
Flexibility and stability of both WT and MTs were further accomplished by measuring root mean square fluctuation (RMSF) at residues level. RMSF is a crucial parameter use to monitor the structural flexibility due to side chain of residues. All MTs and WT showed mean RMSFs in the range of 0.14–0.2, excluding extended loop segment WT, V367F, R408I, G476S, V483A and N501Y MTs, respectively (Fig. 2 A). The extended loop segment present at the C-terminal side displayed higher fluctuation as compared to rest of the protein structures. It comprized ∼26 amino acid long segment (470–495) showed an average RMSFs of ∼0.37, ∼0.70, ∼0.39, ∼0.38, ∼0.32 and ∼0.46 nm for WT, V367F, R408I, G476S, V483A and N501Y MTs, respectively (Fig. 2A). V367F, R408I and N501Y MTs displayed slightly higher RMSFs, whereas V483A MT displayed lower RMSF then WT, indicating that V367F, R408I and N501Y MTs were more flexible and V483A MT was less flexible or more rigid than WT. Residues level inspection showing that Gly476, Ser477, Thr478, Asn481, Gly482 and Val483 remained highly mobile in WT (Fig. 2B) whereas Cys480, Asn481 and Gly482 in V367F MT (Fig. 2C), Ser477 in R408I (Fig. 2D), Cys480 and Asn481 in G476S (Fig. 2E), Ala483 and Glu484 in V483A and Asn481, Gly482 (Fig. 2F), Val483 and Glu484 in N501Y MT (Fig. 2G) residues remained to be highly mobile. RMSF results indicating that fluctuations were more pronounced at the loop region in all MTs and WT.
Fig. 2.
Root mean square fluctuation (RMSF) analysis. (A) RMSF plot at function of amino acid residues, (B) Structure fluctuations of WT, (C) Structure fluctuations of V367F MT, (D) Structure fluctuations of R408I MT, (E) Structure fluctuations of G476S MT, (F) Structure fluctuations of V483A MT and (G) Structure fluctuation of N501Y MT. In 2D RMSF plot, highly fluctuated regions were highlighted and static or mobile regions were shown as blue and red color, respectively. Amino acid residues were depicted in stick mode with 3-letter code. WT, V367F, R408I, G476S, V483A and N501Y MTs were labelled in black, red, green, blue, magenta and yellow color, respectively.
3.3. WT and MTs showed differential structural properties
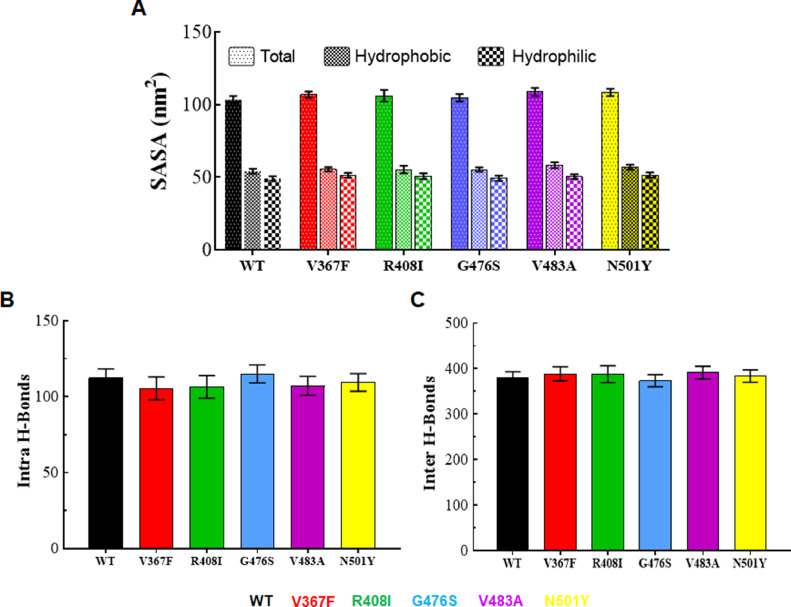
Different structural properties of both WT and MT proteins were assessed through measuring the solvent accessible surface area (SASA), quantitative analysis of intra and inter hydrogen-bonding (H-bond) and qualitative as well as quantitative analyses of secondary structures formation during the entire simulation period. SASA was calculated through gmx sasa module of GROMACS and revealed that total SASA was higher in all MTs as compared to WT. An average SASA (total) values of WT, V367F, R408I, G476S, V483A and N501Y MTs were 103.2, 106.8, 106, 104.6, 108.7 and 108.3 nm2, respectively (Fig. 3 A). Moreover, SASA behaviors remained constant and consistent during an entire simulation period in almost both WT and MTs (Fig. S3). During SASA analysis, hydrophobic remained slightly higher than the hydrophilic SASA in all WT and MT cases. Average values of hydrophobic SASA were lied in the range of 54.2 nm2 for WT (low)–58 nm2 for V483A (high). While hydrophilic SASA values were in the range of 49 nm2 for WT (low)–51.5 nm2 for N501Y (high) (Fig. 3A). This is quite possible because RBD of SpikeS1 formed a core of 7 β-Sheets which is largely hydrophobic in nature.
Fig. 3.
Structural properties analyses of WT and MT proteins. (A) Solvent accessible surface area (SASA) plot, (B) Protein-protein hydrogen bonds (H-bond) and (C) Protein-water hydrogen bonds (H-bond). WT, V367F, R408I, G476S, V483A and N501Y MTs were labelled in black, red, green, blue, magenta and yellow color, respectively.
Structure integrity and interaction capacity of protein determined by various interaction forces out of which hydrogen bond (H-bond) plays a significant role in protein stability and functionality. We calculated both intra (protein-protein) and inter (protein-solvent) H-bond through gmx hbond module of GROMACS. WT, V367F, R408I, G476S, V483A, and N501Y MTs exhibited an average of 112.3, 105.5, 106.4, 115, 107.1, and 109.4 intra H-Bonds, respectively (Fig. 3B), while an average of 378.9, 388.5, 387.8, 373.4, 391.2, and 383.6 H-bonds of WT, V367F, R408I, G476S, V483A and N501Y were formed with the solvent molecule, respectively (Fig. 3C). Moreover, both intra and inter H-bonds remained consistent during an entire simulation period in WT and MT proteins (Fig. S4). During H-bond analysis, we observed that MTs exhibited high number of intra and inter H-bonds than WT except for G476S MT, which formed a lesser number of inter H-bonds. Further, secondary structures such as β-Sheets, helices, β-bridges, bends, turns and coils play an essential role in providing distinct shape and pattern to the protein structures. Each moiety (Sheet, helix, bridge, bend, turn and coil) was further elucidated independently using a dictionary of secondary structure of protein (DSSP) approach and found that helix and β-bridge contents were increased and decreased in MT and WT, respectively. Moreover, all secondary structure moieties remain altered in both WT and MT proteins (Fig. S5).
3.4. Essential motions of WT and MTs
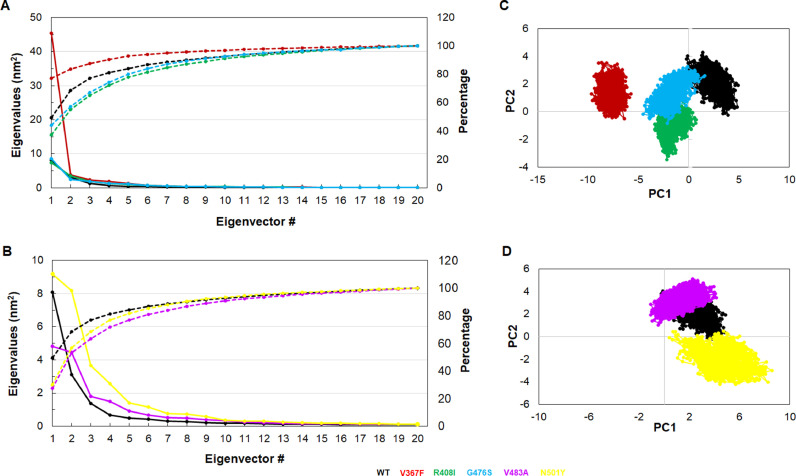
Essential dynamics (ED) which exploited PCA was conducted to elucidate the collective or fundamental motions of WT and MTs. Collective movements are helping to study the conformational changes that existed in the protein, thus reflecting the biological function of that protein. We used stable MD simulation trajectories (125–150 ns) to measure the protein dynamics and sketched the first 20 eigenvectors with their corresponding eigenvalues. Out of which, the first 3 eigenvectors accompanied with maximum motions and their cumulative percentage were 77.04, 87.4, 65.27, 67.33, 63.26 and 68.62% for WT, V367F, R408I, G476S, V483A, and N501Y MTs, respectively (Fig. 4 A and B). Moreover, we found a similar pattern of fluctuations in PC1, PC2, and PC3 as we found during RMSF analysis (Fig. S6). To ensure the biological relevant motions of PCs, we calculated cosine content. We found that the first three PCs of both WT and MT proteins exhibited a low value of cosine (≤0.2), thus indicating that motions were not due to random diffusions (Table 1 ). Conformational sampling of WT and MTs were enquired by plotting and comparing the projection of the first three PCs in phase space in a manner such as PC1 vs PC2, PC1 vs PC3 and PC2 vs PC3. During the projection of PC1 vs PC2 analysis, N501Y occupied a larger sub space whereas the rest of MTs and WT engaged smaller subspaces (Fig. 4C and D). Similar patterns were also observed when projecting the PC1 vs PC3 and PC2 vs PC3 in phase space (Fig. S7). The above results showed that N501Y mutant undergoes large conformational changes.
Fig. 4.
Essential dynamics analysis. (A) Plot of first 20 eigenvectors with associated eigenvalues (solid lines) and their cumulative percentage (dotted lines) of WT, V367F, R408I and G476S MTs, (B) Plot of first 20 eigenvectors with associated eigenvalues (solid lines) and their cumulative percentage (dotted lines) of WT, V483A and N501Y MTs, (C) Projection of PC1 vs PC2 of WT, V367F, R408I and G476S MTs and (D) Projection of PC1 vs PC2 of WT, V483A and N501Y MTs. WT, V367F, R408I, G476S, V483A and N501Y MTs were labelled in black, red, green, blue, magenta and yellow color, respectively.
Table 1.
Cosine content analysis of first two principal components (PC1 and PC2).
| 50 |
100 |
125 |
150 |
|||||
|---|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC1 | PC2 | PC1 | PC2 | PC1 | PC2 | |
| WT | 0.21 | 0.6 | 0.74 | 0.00014 | 0.54 | 0.64 | 0.09 | 0.002 |
| V367F | 0.0063 | 0.02 | 0.84 | 0.0034 | 0.007 | 0.596 | 4.7E-08 | 0.083 |
| R408I | 0.074 | 0.57 | 0.134 | 0.27 | 0.0001 | 0.441 | 0.0053 | 0.075 |
| G476S | 0.144 | 0.859 | 0.132 | 0.66 | 0.0041 | 0.007 | 0.09 | 0.12 |
| V483A | 0.427 | 0.062 | 0.0038 | 0.106 | 0.493 | 0.223 | 0.018 | 0.014 |
| N501Y | 0.021 | 0.514 | 0.682 | 0.054 | 0.00068 | 0.132 | 0.0272 | 0.0128 |
The conformational changes at the structural level were further elucidated by sequentially superimposing the 30 frames of first PC for all WT and MTs (Fig. 5 ). The variations in the motions were mostly found in extended loop regions of MTs as well as WT. Moreover, N501Y MT exhibited additional motions near N-terminal beside the motion at extended loop region (Fig. 5F). Furthermore, the direction and strength of motions were also examined through plotting the porcupine plot of respective PCs (Fig. 5). In porcupine structure, the arrows and length of cone represented direction and magnitude of motions. Different types of motions were exhibited by the extended loop of WT and MT proteins. The upward motions were found in the extended loop of WT and V483A MT (Fig. 5A and E), while downward motions have existed in the loop regions of V367F, R408I and G476S MTs (Fig. 5B–D). Moreover, mixed motions were found in N501Y MT, as downward and upward motions exhibited by the extended loop and rotational motions were found in N-terminal region (Fig. 5F). ED results showed that N501Y MT exhibited a large conformational change at the extended loop and other random moieties of protein, implying that these regions might play a crucial role in acquiring the stable conformation.
Fig. 5.
Major motions analyses of first PC. 30 frames of PC1 were sequentially superimposed and associated motions were depicted in porcupine structures of (A) WT, (B) V367F, (C) R408I, (D) G476S, (E) V483A and (F) N501Y. Intensities and nature of motions were highlighted in dotted circles and arrows in the protein 3D and porcupine structures, respectively.
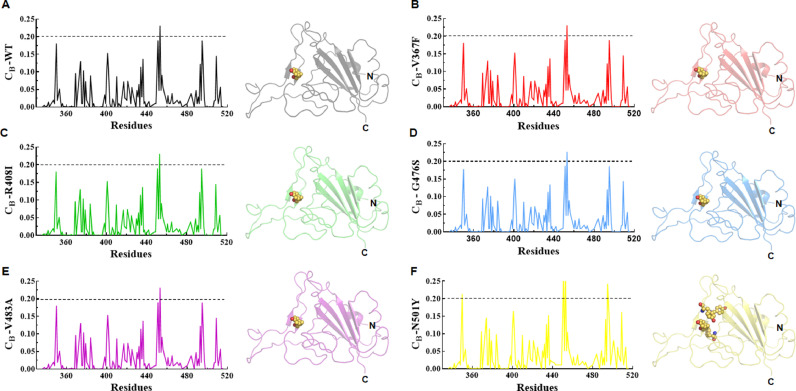
3.5. WT and MTs showed almost similar residues interaction networks
To explore the essential residues involved in the interaction and signalling transduction in the protein, residues interaction network (RIN) analysis was performed for all MTs and WT. MD optimized 3D structures were employed to calculate the centrality and to decipher its connectivity. Here, we computed betweenness centrality (CB), which is a critical centrality based on the shortest route between the nodes and considered the residues with a threshold CB value ≥0.2 (Fig. 6 ). WT and other mutants such as V367F, R408I, G476S, and V483A showed only one residue (Tyrosine453) as par with threshold CB value which is located in the β-sheet region of the protein (Fig. 6A–E). In case of N501Y, four residues, namely Valine 350, Tyrosine451, Tyrosine453 and Tyrosine495 (V350, Y451, Y453 and Y495) were had higher CB values (Fig. 6F). Interestingly, all residues except Y453 lies at the loop region of protein which indeed indicating an important region in the residual signalling of the protein.
Fig. 6.
Network centrality analysis. Betweenness centrality (CB) values of (A) WT, (B) V367F, (C) R408I, (D) G476S, (E) V483A and (F) N501Y. Black dotted line represented the cut off value used to filter the important residues. Right panel of each graph denoted 3D structure displayed in cartoon modes in which important residues shown in sphere.
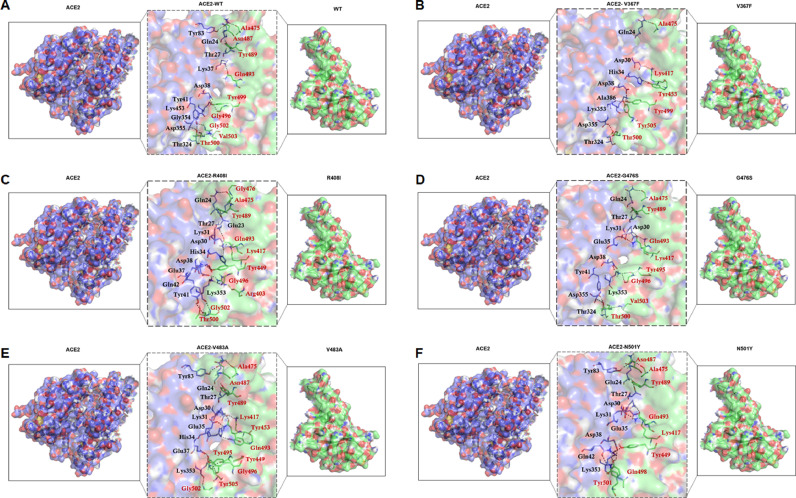
3.6. MTs had altered binding affinities to host protein
Protein–protein (p-p) interaction is a physical association of two or more proteins due to biochemical events driven by many biological forces like Van der Waals, electrostatics and polar interactions. It plays a decisive role in the overall physiology of the cell and its biological process, including signal transduction. Structure based molecular docking approach was widely used to predict the orientation of one protein with respect to other and the key residues involved in the interaction [37]. Here, we employed the HADDOCK server to study the p-p docking of ACE2 (angiotensin-converting enzyme 2) receptor and SpikeS1 RBD of WT and MTs. HADDOCK predicted different clusters for each WT- and MT-protein complexes out of which cluster accompanied larger structures had the lowest HADDOCK score, which indicated the p-p complex of high binding strength. Topmost clusters size of ACE2-WT, -V367F, -R408I, -G476S, -V483A and -N501Y MT complexes were had 168, 169, 127, 175, 176 and 62 structures, respectively (Table 2 ). HADDOCK score of topmost clusters for ACE2-WT, -V367F, -R408I, -G476S, -V483A and -N501Y MT complexes were -122.9 (±3.3), -122 (±2.2), -114.2 (±2), -121.4 (±2.6), -122.1 (±2.3) and -124.4 (±3.1), respectively (Fig. 7 , Table 2). Various energies such as van der Waals, electrostatic, desolvation and restraints violation energies significantly contributed to determining the binding affinities of all WT and MTs p-p interactions (Figs. S8 and S9). During binding energy analysis, electrostatic energy had a major contribution in total binding energy which were -212.3 (±27.8), -185.3 (±29.3), -213.5 (±14.2), -244.6 (±9.7), -214.6 (±18.2) and -210.4 (±20.3) for ACE2-WT, -V367F, -R408I, -G476S, -V483A and -N501Y MT complexes, respectively (Table 2). Additionally, all p-p complexes were stabilized by the ample amount of hydrophilic as well as hydrophobic interactions (Figs. S10 and S11, Table S6). HADDOCK results indicated that there are minor changes in the binding energies of all WT and MTs except R408I. P-p interactions were also predicted by ClusPro and pyDOCKWEB servers (Tables S7 and S8). Furthermore, p-p binding free energies were also verified through the change in binding affinities of ACE2-WT and -MT complexes using mCSM-PPI2 server where positive and negative Delta G represented increase and decrease binding affinities. We have used high scored docking complexes for all WT and MTs obtained from HADDOCK, ClusPro and PyDOCKWEB servers. We found that V367F and V483A MTs showed increased binding affinities while R408I, G476S and N501Y MTs exhibited decreased binding affinities (Table 3 ). The above results indicated that all MTs have differential binding strength to host protein.
Table 2.
Protein-protein (ACE2-RBD) interaction energies analyses.
| Van der Waals energy (stdev.) | Electrostatic energy (stdev.) | Desolvation energy (stdev.) | Restraints violation energy (stdev.) | HADDOCK score (stdev.) | |
|---|---|---|---|---|---|
| WT | −62.9 (±4.8) | −212.3 (±27.8) | −18.4 (±3.2) | 8.3 (±11.8) | −122.9 (±3.3) |
| V367F | −62.8 (±4.4) | −185.3 (±29.3) | −23.8 (±4.2) | 15.7 (±15.1) | −122 (±2.2) |
| R408I | −59.5 (±2.3) | −244.6 (±9.7) | −15.4 (±3.3) | 24.2 (±13.7) | −121.4 (±2.6) |
| G476S | −56.7 (±1.8) | −213.5 (±14.2) | −16.5 (±2.4) | 17.1 (±15.5) | −114.2 (±2.0) |
| V483A | −61.4 (±2.4) | −214.6 (±18.2) | −19.5 (±4.3) | 16.9 (±14.8) | −122.1 (±2.3) |
| N501Y | −59.6 (±2.5) | −210.4 (±20.3) | −24.2 (±3.2) | 14.1 (±13.0) | −124.4 (±3.1) |
Fig. 7.
Protein-protein (ACE2-SpikeS1 RBD) interaction study. (A) ACE2-WT complex, (B) ACE2-V367F complex, (C) ACE2-R408I complex, (D) ACE2-G476S complex, (E) ACE2-V483A complex and (F) ACE2-N501Y complex. ACE2 and SpikeS1 RBD were shown in blue and green surface cartoon modes, respectively. Interacting residues of ACE2 (Black) and SpikeS1 (Red) RBD were represented as stick modes with 3 letter amino acid code.
Table 3.
Binding affinities prediction by mCSM-PPI2 server.
| mCSM-PPI2 prediction of ClusPro P-p docking complexes |
mCSM-PPI2 prediction of pyDockWEB P-p docking complexes |
mCSM-PPI2 prediction of HADDOCK P-p docking complexes |
|
|---|---|---|---|
| Mutation | Predicted Affinity Change (ΔΔG affinity) in kcal/mol | Predicted Affinity Change (ΔΔG affinity) in kcal/mol | Predicted Affinity Change (ΔΔG affinity) in kcal/mol |
| V367F | 0.216 | 0.288 | 0.21 |
| R408I | -0.119 | -0.248 | -0.225 |
| G476S | -0.095 | -0.076 | -0.456 |
| V483A | 0.092 | -0.173 | 0.059 |
| N501Y | -0.219 | -0.116 | -1.075 |
4. Discussion
Viruses, particularly RNA viruses have a marvellous tendency to mutate themselves as compare with DNA viruses, resulting in the rapid emergence of new strains [38]. There are currently thousands of SARS-CoV2 variants reported in which the speed and intensity of spreading some of them (variants) are explained, more than by their genetic differences, by the habits of population, and the effectiveness of epidemiological surveillance policies [39]. SARS-CoV2 is a novel epidemic strain of Betacoronavirus responsible for the widespread viral pandemic COVID-19 [40]. The virus gets into the cell with the help of spike glycoprotein (S1) present on the surface of the virus envelope which interacts with the ACE2 receptor of host cell [7]. Receptor binding domain (RBD) of SpikeS1 protein interacts with ACE2 receptors and are more prone to mutations resulting in the formation of new strains and also help the viruses to escape from different therapies [15]. The RBD consists of a core region of β-sheets and an extended loop region known as receptor binding motifs that provide a binding surface to the ACE2 protein [16]. Before developing a specific and effective drug, one must understand the molecular interaction of ACE2 and SpikeS1 protein of different strains. Therefore, the present study has critically analyzed the effect of various mutations on the structural stability and dynamics of RBD of SpikeS1 protein by manifesting different MTs or strains of SARS-CoV2 and assessed the binding affinities with host (human) ACE2 receptor.
The tertiary structure of ACE2-SpikeS1 RBD complex was taken from PDB and used as a template for creating various SARS-CoV2 mutants. Qualities of generated mutant models were verified by inspecting the stereochemical and geometrical properties through different model validation tools and found that all MTs (V367F, R408I, G476S, V483A and N501Y) had good profiles of Ramachandran plots, acceptable scores of ProSA and QMEAN, similar to WT protein, implying that all MTs along with WT had better stereochemical and geometrical properties. To explore the structural and functional properties, the optimized RBD MTs were subjected to MD simulations of 150 ns simulation time. MD simulation is a promising technique to study the stability and dynamics of macromolecules at an atomic level in a realistic manner, which is hard to achieve through experimental methods [41,42]. Different parameters such as RMSD, RMSF and Rg were measured to monitor the stability and dynamics of WT and MT proteins. RMSD results suggested that WT as well as MT proteins displayed consistent behavior with higher RMSDs were found in V367F, R408I and V483A MTs as compared to WT, G476S and N501Y MTs. RMSF analysis was performed to study the residue and structural level alterations in WT and MTs and found residues like Gly476, Ser477, Thr478, Asn481, Gly482 and Val483 were highly fluctuated and located at loop regions of almost all MTs and WT [43]. Loops are essential moieties of protein that are highly mobile and act as a crucial site for ligand interactions and help in acquiring the stable conformation of proteins [44]. Hence, the mobility of residues in the loop regions in WT and MTs help in the interactions of RBD to partner protein such as ACE2. Previous study at microsecond MD simulation suggested that loop region of receptor binding motif persistently involved in ACE interaction and residues lied in this region are critical for mediating the entrance of SARSCoV-2 into host cells [45,46].
Next, we have analyzed the structural properties of all MTs along with WT by measuring SASA, H-bonds and secondary structure units. The core region of protein mainly consists of hydrophobic residues that act as a driving force during protein folding. The solvent interaction intensity of amino acid and the core of protein is proportional to the total surface area exposed to these environments [47]. Therefore, SASA of WT and MTs were measured to monitor the surface area available for the surrounding environment and found that all MTs displayed higher SASA then WT and hydrophobic SASA contributed slightly more in comparison to hydrophilic SASA as RBD of protein mostly consist of core followed by loop regions. Hydrogen bonding (H-bond) is an important parameter for providing stability and strength of protein-ligand interaction properties of a particular protein. The stability and protein-ligand interaction capacity were measured by calculating the intra (protein-protein) and inter (protein-solvent) H-bonds [48]. H-bond results suggested that no significant changes in overall intra and inter H-bonds were existed in all MTs compared to WT. Furthermore, secondary structure analyses of all MTs and WT indicated that alterations in the random moieties of protein compromized with changing in α-helix and β-sheet contents.
ED, a typical application of PCA, which is used to extract the biological relevant motions by utilising the MD simulation trajectories [49,50]. Moreover, essential dynamics helps enable to study the free energy landscape of different MTs in apo and complex forms [51]. Equilibrated MD trajectories were harnessed for constructing the covariance matrix and series of eigenvectors with associated eigenvalues were plotted for all MTs and WT. The first three eigenvectors were used to study the conformational subspace of MTs and WT protein as they are mostly encompassing the major motions. ED results suggested that, except for N501Y MT, all other MTs were had similar conformational subspace as with WT, while N501Y MT covered larger conformational space. Moreover, the structure level inspection from the first eigenvector (PC1) revealed that the higher motions were accompanied by extended loop region of the protein. The biological function of a protein is characterizing by constituent residues and their chemical properties. These chemical versatilities of amino acids determine the overall topology of protein and its behavior in the surrounding environment. Different residues in the protein interact and help in the flow of signal, which is better elucidated by the residues interaction network (RIN) [52]. In this analysis, an entire protein considered as a network in which residues and interactions are plotted in the form of nodes and edges, respectively [53]. Three centralities were computed out of which betweenness centrality (CB) is most important concerning to protein function [42]. RIN results suggested that Tyrosine453 was an important residue in WT and V367F, R408I, G476S, and V483A MTs, while more than one amino acid had role in signalling in N501Y MT. The above residues are mainly located on or near the 5th and 6th β-sheets of RBD, demonstrating that this region might be crucial for the functioning of WT and MT proteins. Previous study showed that mutation shorten the signalling path between the residues in RBD that makes the residues in close proximities and impart strong signalling which may lead to effective opening or closing of loop regions [54].
SARS-CoV2 invades host cells through the interaction of RBD of SpikeS1 unit to the host ACE2 receptor. Qualitatively, the infectivity of diverse SARS-CoV2 strains in the host is proportional to the binding affinity of SARS-CoV2 SpikeS1 RBD of each strain to the ACE2 receptor of host cells [55]. Therefore, the assessment of binding affinity of different MTs to host cells is vital for understanding the infectiousness of SARS-CoV2. The binding energies of WT and MTs RBD to ACE2 was elucidated by the HADDOCK, ClusPro and pyDOCKWEB protein-protein (p-p) docking server and binding affinities were calculated through mCSM-PPI2 server. P-p interaction results suggested that V367F and V483A MTs showed increased binding affinities predicted while R408I, G476S and N501Y MTs exhibited decreased binding affinities. Binding affinity analysis indicated that V367F and V483A MTs binds to host ACE2 protein with maximum binding strength, thus contributing highly infectious activity.
Proteins are having dynamic personalities and are central to the molecular function of a cell. Any change in amino acid residue of proteins led to the changing in overall topology and hence affect their conformations and thus functions [56]. The molecular and functional consequences of changes in amino acids are better observed in the current pandemic (COVID-19) as the point mutations in the SpikeS1 RBD of SARS-CoV2 result in the generation of thousands of variants thus, led to the emergence of various strains [57]. The alterations in amino acids showed both local and the global impact on the topology of protein and in turn, altered the interacting residue, which ultimately affected the binding affinity of two partner proteins (ACE2-SpikeS1 RBD). In V367F MT, a nonpolar hydrophobic amino acid has aliphatic side chain (Valine) is mutated to an aromatic hydrophobic residue (Phenylalanine) that enhanced the structural stability of the spike protein, thus facilitating more efficient binding to the host ACE2 receptor as also shown by pyDOCKWEB server (Table S8). In case of R408I MT, there is change in overall charge of protein residue (Arginine), which is mutated to the residues having a neutral charge (Isoleucine). This also affected overall topology of protein as mutated residue have a smaller size than WT residue, resulting in the decreasing of overall binding affinity as reported during p-p interaction. In G476S MT, Glycine of WT residues is mutated to Serine which affects the overall flexibility of protein as Glycine is more flexible than Serine. This led to a decrease in the stability and binding affinity of G476S MT, as noticed during MD simulation and p-p docking. In V483A MT Valine, a large hydrophobic protein residue is mutated to Alanine, a small size hydrophobic residue that alters the overall dimension of the protein, thus help in increasing the binding affinity. And, in case of N501Y MT, where a substitution of Asparagine to Tyrosine was found in the binding motif of SpikeS1 RBD, which has increased binding affinity with its protein partner as shown by pyDOCKWEB server and ultimately leads to a stable virus-host relationship (Table S8). Additionally, binding affinity of receptor-ligand was were enhanced by bending angle, signifying more inclination of RBD towards ACE2 complex that facilitated more tightly holding in the binding site and thus strengthened the affinity as reported previously [58]. Further, this mutation also assists the virus in evading antibody neutralization by changing the configuration of epitope site [59].
The present scenario of global pandemic, where the world keeps an eye towards the vaccine at utmost level. In this regard, the computational approaches have the potential to unravel the mechanistic behavior of RBD MTs and their interactions with ACE receptor at an atomic level which aid in guiding the drug development process. However, certain limitations are associated with the current computational study, which may be overcome by experimental methods. Therefore, experimental methods are highly urged to support or validate the current findings. Nevertheless, our study provides computational evidence of the strength of highly infectious strains, which would help in design a more specific drug to combat the current pandemic.
Statement of ethical approval
Not applicable
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not for profit sectors.
CRediT authorship contribution statement
Rakesh Kumar: Conceptualization, Visualization, Methodology, Formal analysis, Writing – original draft, Writing – review & editing. Rahul Kumar: Methodology, Formal analysis, Writing – original draft, Writing – review & editing. Harsh Goel: Writing – original draft, Writing – review & editing. Pranay Tanwar: Conceptualization, Visualization, Writing – review & editing.
Declaration of Competing Interest
The authors have no conflict of interest to declare
Acknowledgments
Rakesh and Rahul thank Indian Council of Medical Research and University Grant Commission, respectively for financial support. H.G. acknowledged CSIR (Council of Scientific and Industrial Research), India for financial assistance.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cmpb.2021.106594.
Appendix. Supplementary materials
References
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:30185–30189. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roser M., Ritchie H., Ortiz-Ospina E., Hasell J. Published online at OurWorldInData.org; 2020. “Coronavirus Pandemic (COVID-19)”. Retrieved from: 'https://ourworldindata.org/coronavirus' [Online Resource] [Google Scholar]
- 3.Bontempi E. The Europe second wave of COVID-19 infection and the Italy "strange" situation. Environ. Res. 2021;193 doi: 10.1016/j.envres.2020.110476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young B.E., Fong S.W., Chan Y.H., Mak T.M., Ang L.W., Anderson D.E., Lee C.Y., Amrun S.N., Lee B., Goh Y.S., Su Y.C.F., Wei W.E., Kalimuddin S., Chai L.Y.A., Pada S., Tan S.Y., Sun L., Parthasarathy P., Chen Y.Y.C., Barkham T., Lin R.T.P., Maurer-Stroh S., Leo Y.S., Wang L.F., Renia L., Lee V.J., Smith G.J.D., Lye D.C., Ng L.F.P. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396:603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menachery V.D., Yount B.L., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., Swanstrom J., Sheahan T.P., Pickles R.J., Corti D., Randell S.H., Lanzavecchia A., Marasco W.A., Baric R.S. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U.S.A. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E.E., Bhattacharya T., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield, Genomics Group, McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;184:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J., Petitjean S.J.L., Koehler M., Zhang Q., Dumitru A.C., Chen W., Derclaye S., Vincent S.P., Soumillion P., Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saha P., Banerjee A.K., Tripathi P.P., Srivastava A.K., Ray U. A virus that has gone viral: amino acid mutation in S protein of Indian isolate of Coronavirus COVID-19 might impact receptor binding, and thus, infectivity. Biosci. Rep. 2020;40 doi: 10.1042/BSR20201312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang R., Chen J., Gao K., Hozumi Y., Yin C., Wei G.W. Analysis of SARS-CoV-2 mutations in the United States suggests presence of four substrains and novel variants. Commun. Biol. 2021;4:228. doi: 10.1038/s42003-021-01754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wise J. Covid-19: new coronavirus variant is identified in UK. BMJ. 2020;371:m4857. doi: 10.1136/bmj.m4857. [DOI] [PubMed] [Google Scholar]
- 14.H. Tegally, E. Wilkinson, M. Giovanetti, A. Iranzadeh, V. Fonseca, J. Giandhari, D. Doolabh, S. Pillay, E.J. San, N. Msomi, K. Mlisana, A.V. Gottberg, S. Walaza, M. Allam, A. Ismail, T. Mohale, A.J. Glass, S. Engelbrecht, G.V. Zyl, W. Preiser, F. Petruccione, A. Sigal, D. Hardie, G. Marais, M. Hsiao, S. Korsman, M.A. Davies, L. Tyers, I. Mudau, D. York, C. Maslo, D. Goedhals, S. Abrahams, O.L. Akingba, A.A. Dehkordi, A. Godzik, C.K. Wibmer, B.T. Sewell, J. Lourenco, L.C.J. Alcantara, S.L.K. Pond, S. Weaver, D. Martin, R.J. Lessells, J.N. Bhiman, C. Williamson, T.D. Oliveira, Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa, 2020, medRxiv 2020.12.21.20248640.
- 15.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 16.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose P.W., Beran B., Bi C., Bluhm W.F., Dimitropoulos D., Goodsell D.S., Prlic A., Quesada M., Quinn G.B., Westbrook J.D., Young J., Yukich B., Zardecki C., Berman H.M., Bourne P.E. The RCSB protein data bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:392–401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ningombam S.S., Kumar R., Tanwar P. Mutant strains of SARS-CoV-2 are more prone to infect obese patient: a review. Wien. Klin. Wochenschr. 2021;133:383–392. doi: 10.1007/s00508-021-01819-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 20.Luthy R., Bowie J.U., Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 21.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:407–410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benkert P., Tosatto S.C., Schomburg D. QMEAN: a comprehensive scoring function for model quality assessment. Proteins. 2008;71:261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- 23.Oostenbrink C., Villa A., Mark A.E., van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 25.Amadei A., Linssen A.B., Berendsen H.J. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 26.Hess B. Similarities between principal components of protein dynamics and random diffusion. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Top. 2020;62:8438–8448. doi: 10.1103/physreve.62.8438. [DOI] [PubMed] [Google Scholar]
- 27.Kumar R., Saran S. Structure, molecular dynamics simulation, and docking studies of Dictyostelium discoideum and human STRAPs. J. Cell. Biochem. 2018;119:7177–7191. doi: 10.1002/jcb.26840. [DOI] [PubMed] [Google Scholar]
- 28.Chakrabarty B., Parekh N. NAPS: network analysis of protein structures. Nucleic Acids Res. 2016;44:375–382. doi: 10.1093/nar/gkw383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Zundert G.C.P., Rodrigues J., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A. The HADDOCK2.2 web server: user-friendly integrative modelling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Kumar R., Mukherjee P.K. Trichoderma virens Bys1 may competitively inhibit its own effector protein Alt a 1 to stabilize the symbiotic relationship with plant evidence from docking and simulation studies. 3 Biotech. 2021;11:144. doi: 10.1007/s13205-021-02652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comeau S.R., Gatchell D.W., Vajda S., Camacho C.J. ClusPro: a fully automated algorithm for protein–protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiménez-García B., Pons C., Fernández-Recio J. pyDockWEB: a web server for rigid-body protein–protein docking using electrostatics and desolvation scoring. Bioinformatics. 2013;29:1698–1699. doi: 10.1093/bioinformatics/btt262. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigues C.H.M., Myung Y., Pires D.E.V., Ascher D.B. mCSM-PPI2: predicting the effects of mutations on protein-protein interactions. Nucleic Acids Res. 2019;47:338–344. doi: 10.1093/nar/gkz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng J., Randall A., Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- 36.Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:306–310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maurya R., Kumar R., Saran S. AMPKα promotes basal autophagy induction in Dictyostelium discoideum. J. Cell. Physiol. 2020;235:4941–4953. doi: 10.1002/jcp.29373. [DOI] [PubMed] [Google Scholar]
- 38.Elena S.F., Sanjuan R. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 2005;79:11555–11558. doi: 10.1128/JVI.79.18.11555-11558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodcroft E.B., Zuber M., Nadeau S., Vaughan T.G., Crawford K.H.D., Althaus C.L., Reichmuth M.L., Bowen J.E., Walls A.C., Corti D., Bloom J.D., Veesler D., Mateo D., Hernando A., Comas I., González-Candelas F., SeqCOVID-SPAIN consortium, Stadler T., Neher R.A. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature. 2021;595:707–712. doi: 10.1038/s41586-021-03677-y. [DOI] [PubMed] [Google Scholar]
- 40.Wong N.A., Saier M.H. The SARS-coronavirus infection cycle: a survey of viral membrane proteins, their functional interactions and pathogenesis. Int. J. Mol. Sci. 2021;22:1308. doi: 10.3390/ijms22031308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar R., Maurya R., Saran S. Identification of novel inhibitors of the translationally controlled tumor protein (TCTP): insights from molecular dynamics. Mol. Biosyst. 2017;13:510–524. doi: 10.1039/c6mb00850j. [DOI] [PubMed] [Google Scholar]
- 42.Kumar R., Kumar R., Tanwar P., Deo S.V.S., Mathur S., Agarwal U., Hussain S. Structural and conformational changes induced by missense variants in the zinc finger domains of GATA3 involved in breast cancer. RSC Adv. 2020;10:39640–39653. doi: 10.1039/d0ra07786k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yogurtcu O.N., Gur M., Erman B. Statistical thermodynamics of residue fluctuations in native proteins. J. Chem. Phys. 2009;130 doi: 10.1063/1.3078517. 095103. [DOI] [PubMed] [Google Scholar]
- 44.Baiesi M., Orlandini E., Seno F., Trovato A. Sequence and structural patterns detected in entangled proteins reveal the importance of co-translational folding. Sci. Rep. 2019;9:8426. doi: 10.1038/s41598-019-44928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spinello A., Saltalamacchia A., Magistrato A. Is the rigidity of SARS-CoV-2 spike receptor-binding motif the hallmark for its enhanced infectivity? Insights from all-atom simulations. J. Phys. Chem. Lett. 2020;11:4785–4790. doi: 10.1021/acs.jpclett.0c01148. [DOI] [PubMed] [Google Scholar]
- 46.Serapian S.A., Marchetti F., Triveri A., Morra G., Meli M., Moroni E., Sautto G.A., Rasola A., Colombo G. The answer lies in the energy: how simple atomistic molecular dynamics simulations may hold the key to epitope prediction on the fully glycosylated SARS-CoV-2 spike protein. J. Phys. Chem. Lett. 2020;11:8084–8093. doi: 10.1021/acs.jpclett.0c02341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pace C.N., Fu H., Lee Fryar K., Landua J., Trevino S.R., Schell D., Thurlkill R.L., Imura S., Scholtz J.M., Gajiwala K., Sevcik J., Urbanikova L., Myers J.K., Takano K., Hebert E.J., Shirley B.A., Grimsley G.R. Contribution of hydrogen bonds to protein stability. Protein Sci. 2014;23:652–661. doi: 10.1002/pro.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar R., Saran S. Comparative modelling unravels the structural features of eukaryotic TCTP implicated in its multifunctional properties: an in-silico approach. J. Mol. Model. 2021;27:20. doi: 10.1007/s00894-020-04630-y. [DOI] [PubMed] [Google Scholar]
- 49.Berendsen H.J., Hayward S. Collective protein dynamics in relation to function. Curr. Opin. Struct. Biol. 2000;10:165–169. doi: 10.1016/s0959-440x(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 50.Skjaerven L., Hollup S.M., Reuter N. Normal mode analysis for proteins. J. Mol. Struct. 2009;898:42–48. [Google Scholar]
- 51.Pontiggia F., Colombo G., Micheletti C., Orland H. Anharmonicity and self-similarity of the free energy landscape of protein G. Phys. Rev. Lett. 2007;98 doi: 10.1103/PhysRevLett.98.048102. [DOI] [PubMed] [Google Scholar]
- 52.Joshi K., Kaur S., Kumar R. Cytochrome P450 2C19 gene polymorphisms (CYP2C19* 2 and CYP2C19* 3) in chronic myeloid leukemia patients: in vitro and in silico studies. J. Biomol. Struct. Dyn. 2021:1–14. doi: 10.1080/07391102.2021.1929491. [DOI] [PubMed] [Google Scholar]
- 53.Przulj N., Wigle D.A., Jurisica I. Functional topology in a network of protein interactions. Bioinformatics. 2004;20:340–348. doi: 10.1093/bioinformatics/btg415. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y., Liu M., Gao J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc. Natl. Acad. Sci. U. S. A. 2020;117:13967–13974. doi: 10.1073/pnas.2008209117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mittal A., Manjunath K., Ranjan R.K., Kaushik S., Kumar S., Verma V. COVID-19 pandemic: insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLOS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar R. Mutations in passive residues modulate 3D-structure of NDM (New Delhi metallo-β-lactamase) protein that endue in drug resistance: a MD simulation approach. J. Biomol. Struct. Dyn. 2021:1–17. doi: 10.1080/07391102.2021.1930165. [DOI] [PubMed] [Google Scholar]
- 57.Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spinello A., Saltalamacchia A., Borišek J., Magistrato A. Allosteric cross-talk among spike's receptor-binding domain mutations of the SARS-CoV-2 South African variant triggers an effective hijacking of human cell receptor. J. Phys. Chem. Lett. 2021;12:5987–5993. doi: 10.1021/acs.jpclett.1c01415. [DOI] [PubMed] [Google Scholar]
- 59.Triveri A., Serapian S.A., Marchetti F., Doria F., Pavoni S., Cinquini F., Moroni E., Rasola A., Frigerio F., Colombo G. SARS-CoV-2 spike protein mutations and escape from antibodies: a computational model of Epitope loss in variants of concern. J. Chem. Inf. Model. 2021;61:4687–4700. doi: 10.1021/acs.jcim.1c00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.