Figure 1.
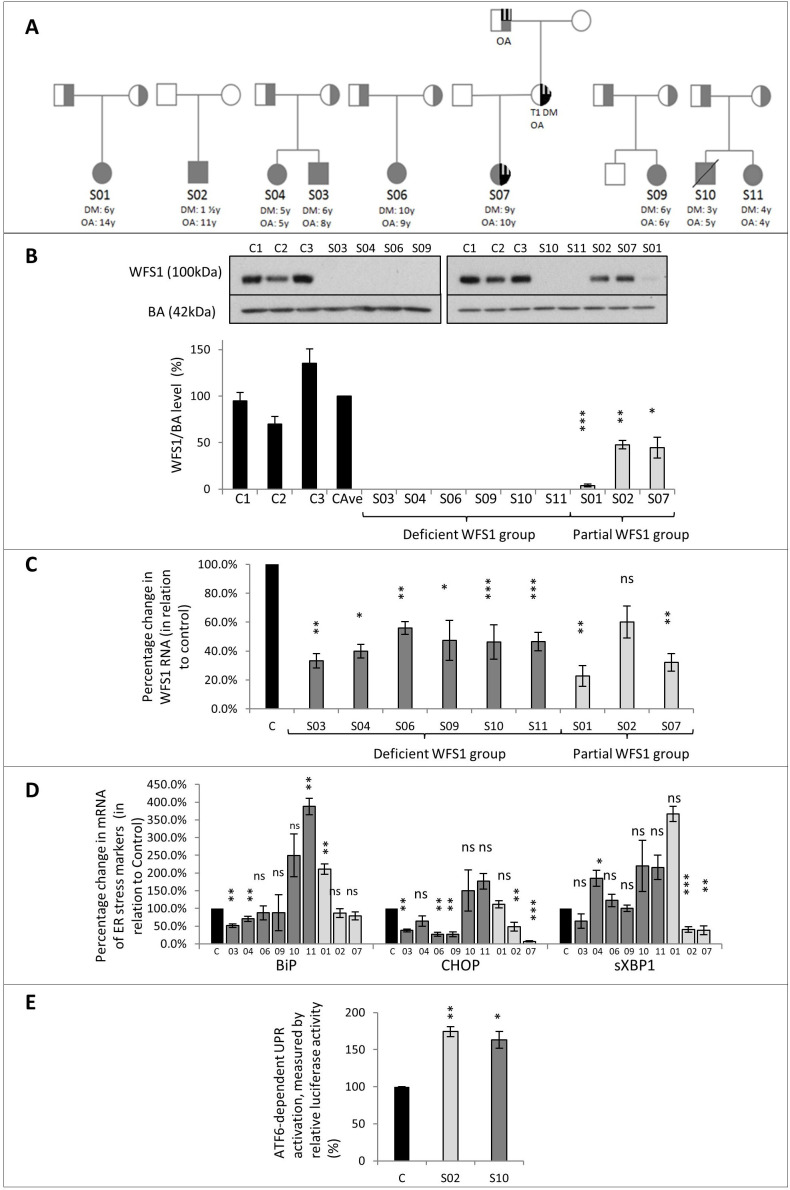
Family pedigrees and functional data.(A) Pedigrees of the 7 families reported in this study. All patients included in the study marked in grey. The age of onset of diabetes mellitus (DM) and optic atrophy (OA) stated. S02 had a de-novo mutation. For patient S07, OPA1 and WFS1 variants were found in all three generations in this family: S07’s maternal grandfather had isolated OA; S07’s mother had isolated OA and Type 1 DM. OA and DM in S07’s family represented by quarter stripes (OA) and quarter black (DM). All other patients inherited recessive alleles from each parent.(B) Immunoblotting images and corresponding bar chart with standard error bars showing levels of WFS1 protein. WFS1 and beta-actin (BA) protein levels measured in fibroblast from patients with WFS and healthy controls. C1, C2, C3 = healthy controls; CAve: average of controls. WFS1 levels for CAve=100%. WFS1 protein was undetectable in patients: S03, S04, S06, S09, S10 and S11. WFS1 protein was reduced in S01, S02 and S07 by 96.2%, 53.3%, and 55.4% respectively in comparison to CAve. Analysis by Student's T-test.(C) Bar chart with standard error bars showing quantitative PCR analysis of WFS1 mRNA, as percentage change when standardised with control. C=control. (n= 4) Analysis by Student's T-test.(D) Bar chart with standard error bars showing quantitative PCR analysis of ER stress marker mRNA: BiP, CHOP and sXBP1, as percentage change, when standardised with control (C). (n=4). Dark grey bars indicate patient in the deficient WFS1 protein group, and the light grey bar indicates the patient is in the partial WFS1 protein group. Analysis by Student's T-test.(E) Bar chart with standard error bars showing quantification of ATF6-dependent UPR activation by ERSE luciferase reporters, for SO2 and S10, as a percentage change compared from control (C). (n=4) Analysis by Student's T-test.ns: P >0.05; * P≤0.05; ** P ≤0.01; *** P≤0.001 compared with control samples