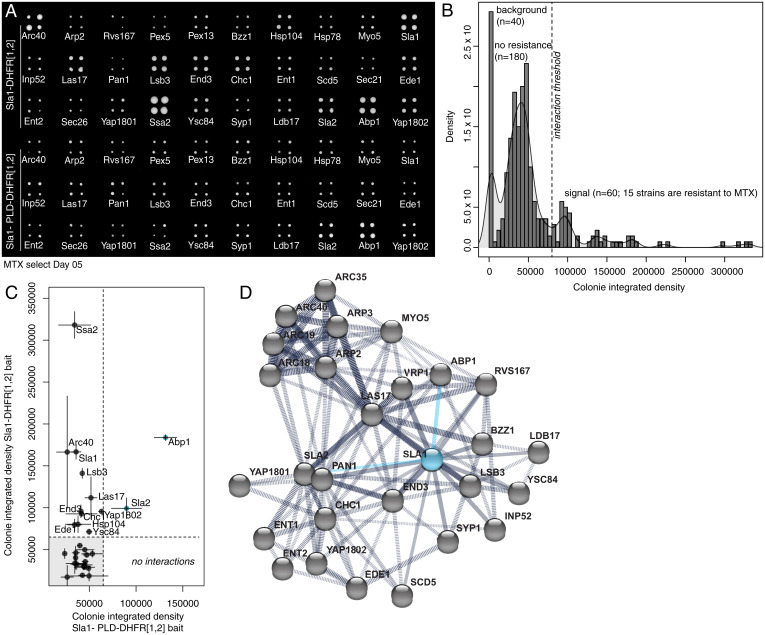
Fig. 4.
Proteins can partition into endocytic puncta through a network for interactions coordinated by PLDs. (A) DHFR PCA and methotrexate (MTX) selection plate after 5 d of growth shows that the PLD deletion mutant of Sla1 loses all but two interactions of wild-type Sla1. (B) Density (number of colonies with given total pixel intensity) versus colony-integrated growth (total pixel intensity) after 5 d of MTX selection for DHFR reconstitution. Peaks are labeled with their corresponding measurements for plate background (n = 40 empty spots), colonies with no resistance to MTX (n = 180 colonies; 45 strains), and colonies that grow on MTX (n = 60 colonies; 15 strains) above a selected integrated density threshold. We detected 13 interactions for full-length Sla1 protein. (C) Colony-integrated growth (total pixel intensity) after 5 d of MTX selection for DHFR reconstitution with wild-type Sla1 versus colony integrated growth (total pixel intensity) after 5 d of MTX selection for DHFR reconstitution with PLD deletion mutant of Sla1. Compared to wild-type Sla1 interactions, we detect two interactions (blue points) for the mutant Sla1 protein that do not contain the PLD. (D) Protein interaction network from STRING (version 11.0) for the selected subarray of 30 potential interactors (gray dots) of Sla1 (blue dot). We represent the 13 direct Sla1 interactions (solid black lines) and DHFR PCA interactions with Apb1 and Sla2 that are preserved with the PLD deletion mutant of Sla1 (solid blue lines) among all the protein–protein interactions (dashed black lines) determined experimentally.