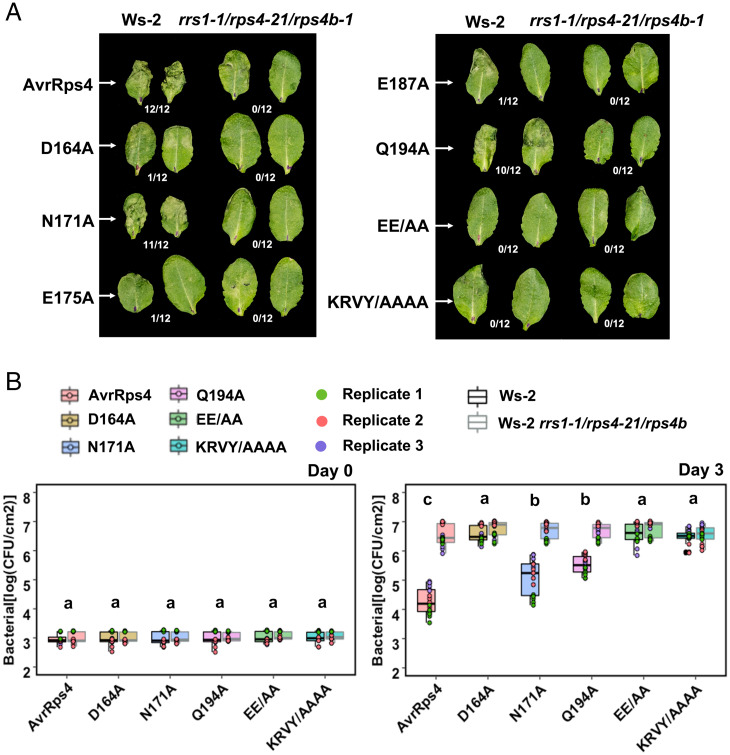
Fig. 5.
Structure-guided mutants of AvrRps4 compromise RRS1-R/RPS4–dependent recognition specificities and restriction of bacterial growth in Arabidopsis. (A) HR assay in different Arabidopsis accessions using P. fluorescens Pf0-1 secreting AvrRps4 wild type and structure-guided mutants. Constructs were delivered to the Arabidopsis Ws-2 and rrs1-1/rps4-21/rps4b-1 knockout background and HR was recorded 20 h post infiltration. Fraction refers to the number of leaves showing HR of 12 randomly inoculated leaves. This experiment was repeated at least three times with similar results. (B) In planta bacterial growth assays of Pto DC3000 secreting AvrRps4 wild type and mutant constructs. Bacterial suspensions with OD600 = 0.001 were pressure-infiltrated into the leaves of 4- to 5-wk-old Arabidopsis plants. Values are plotted from three independent experiments (denoted in different colors). Statistical significance of the values was calculated by one-way ANOVA followed by post hoc Tukey honestly significant difference analysis. Letters above the data points denote significant differences (P < 0.05). A detailed statistical summary can be found in SI Appendix, Table S5. CFU, colony forming unit.