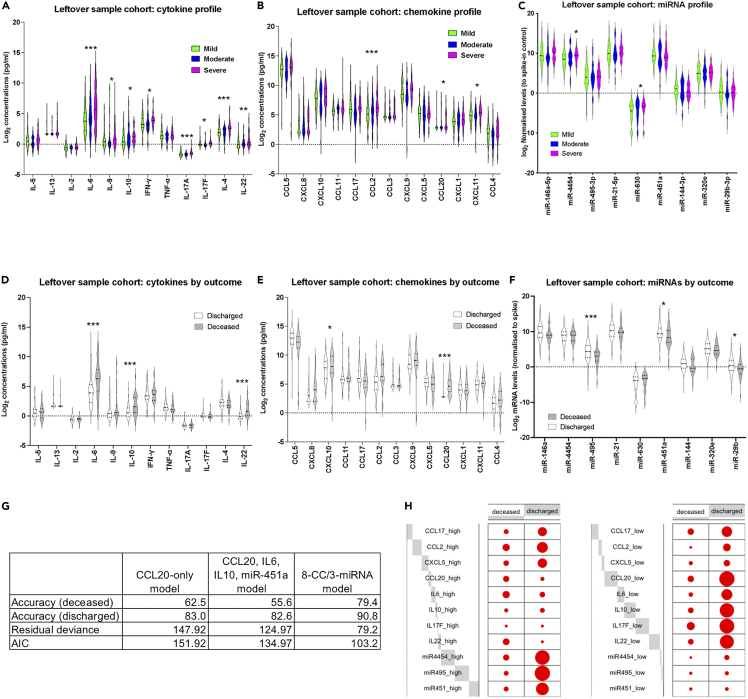
Figure 7.
miRNA/CC correlates of fatal COVID-19 identified in a leftover blood sample cohort
(A) Violin plots of measured cytokines in mild (green), moderate (blue), and severe (magenta) samples, in the leftover blood sample cohort. Stars indicate significance for the comparison of severe against mild and moderate as follows: ∗ adjusted p <0.05, ∗∗ adjusted p <0.01, ∗∗∗ adjusted p <0.005.
(B) As in A, but for chemokine levels.
(C) As in A, but for miRNA levels.
(D) Violin plots of measured cytokines in samples from discharged (white) and deceased (gray) patients, in the leftover blood sample cohort. Stars indicate significance for the comparison of severe against mild and moderate as follows: ∗ adjusted p <0.05, ∗∗ adjusted p <0.01, ∗∗∗ adjusted p <0.005.
(E) As in D, but for chemokine levels.
(F) As in D, but for miRNA levels.
(G) Results of forward regression models including accuracy for deceased and discharged patients, residual deviance, and AIC. Results are shown for the CCL20-only model, the CCL20/IL6/IL10/miR-451a model, and the 3 miRNAs/11 CC models (see Table S11).
(H) Balloon plots for the parameters included in the 3 miRNAs/11 CC model. The size of each dot reflects the number of patients in the deceased or discharged group with high (left panel), or low (right panel) levels of the shown CCs and miRNAs. Gray blocks indicate the proportion of the total counts (high or low) each variable accounts for. See STAR methods for definition of high and low values.