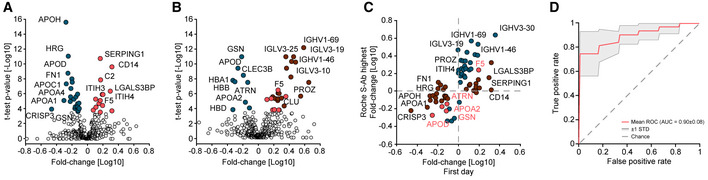
Figure 2. Serum proteome differences of COVID‐19 patients and SARS‐CoV‐2 PCR‐negative controls with COVID‐19‐like symptoms.
-
AVolcano plot comparing the serum proteomes of 31 COVID‐19 patients on the first day of sampling to those of the 262 PCR‐negative controls. Significantly up‐regulated proteins in COVID‐19 positive patients are highlighted in red and down‐regulated proteins in blue. Highlighted proteins are significant after multiple hypothesis testing. The log10 fold‐change in protein levels is represented on the x‐axis and the −log10 t‐test P‐value on the y‐axis. Examples of significantly altered proteins are labeled.
-
BVolcano plot comparing the serum proteomes in samples from COVID‐19 patients at the time point of highest Roche S‐Ab levels to PCR‐negative controls. Significantly up‐regulated proteins in COVID‐19 positive patients are highlighted in red and down‐regulated proteins in blue. Significantly up‐regulated immunoglobulin regions are highlighted in dark red. Examples of significantly altered proteins are labeled.
-
CScatter plot of protein fold‐changes in (A) vs. those in (B). Significant proteins of (A) are highlighted dark red, those of (B) in blue, and significant in both in bright red. Examples of significantly altered proteins are labeled.
-
DROC curve to classify whether a sample was obtained from a COVID‐19 positive or a PCR‐negative control patient. The mean ROC curve is displayed in red and ± 1 standard deviations are illustrated in gray. The model achieved an area under the curve (AUC) of 0.90.
