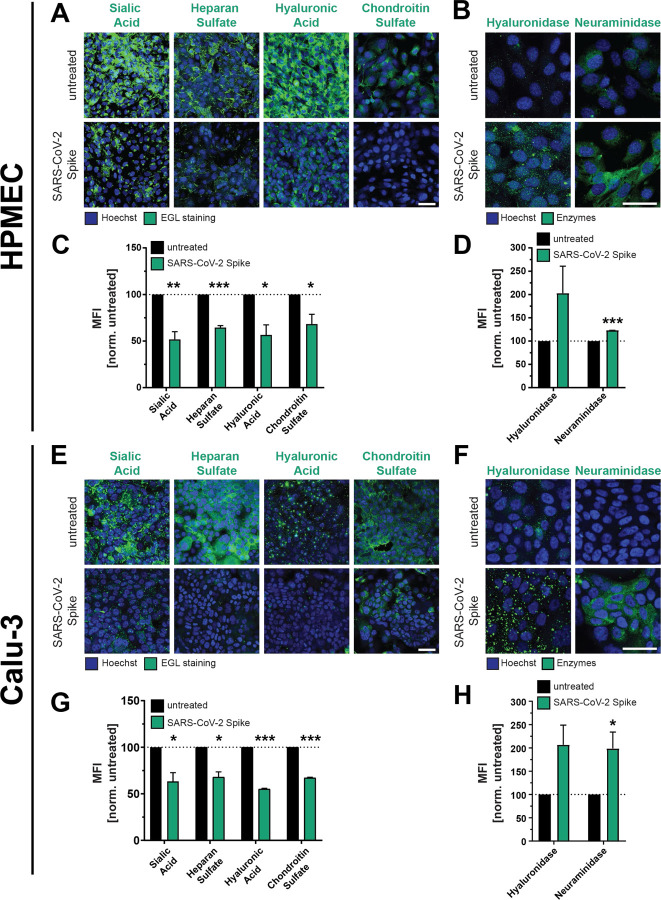
Figure 2. SARS-CoV-2 S facilitates disruption of the endothelial and epithelial glycocalyx layer.
(A) An immunofluorescence microscopy-based EGL disruption assay measuring levels of the indicated glycans on the surface of HPMEC. After 24 h of SARS-CoV-2 S (10 μg/mL) treatment, cells were fixed then stained without permeabilization. Displayed are representative images from n=3 biological replicates. (B) Same as A, but cells were permeabilized before staining for the indicated EGL disrupting enzymes. Displayed are representative images from n=3 biological replicates. (C) Quantification of A. (D) Quantification of B. (E) Same as A but measuring EGL disruption of Calu-3 cell monolayers. Displayed are representative images from n=3 biological replicates. (F) Same as B but measuring expression of EGL-disrupting enzymes in Calu-3 cells. Displayed are representative images from n=3 biological replicates. (G) Quantification of E. (H) Quantification of F. For all images, nuclei were probed with Hoechst in blue and the indicated glycans in green with scale bars at 100 μm. Dotted lines are the normalized untreated control conditions. MFI is mean fluorescence intensity. All data are plotted as mean +/− SEM with * p<0.05, ** p<0.01, *** p<0.001, and n.s. p>0.05 by unpaired t-test.