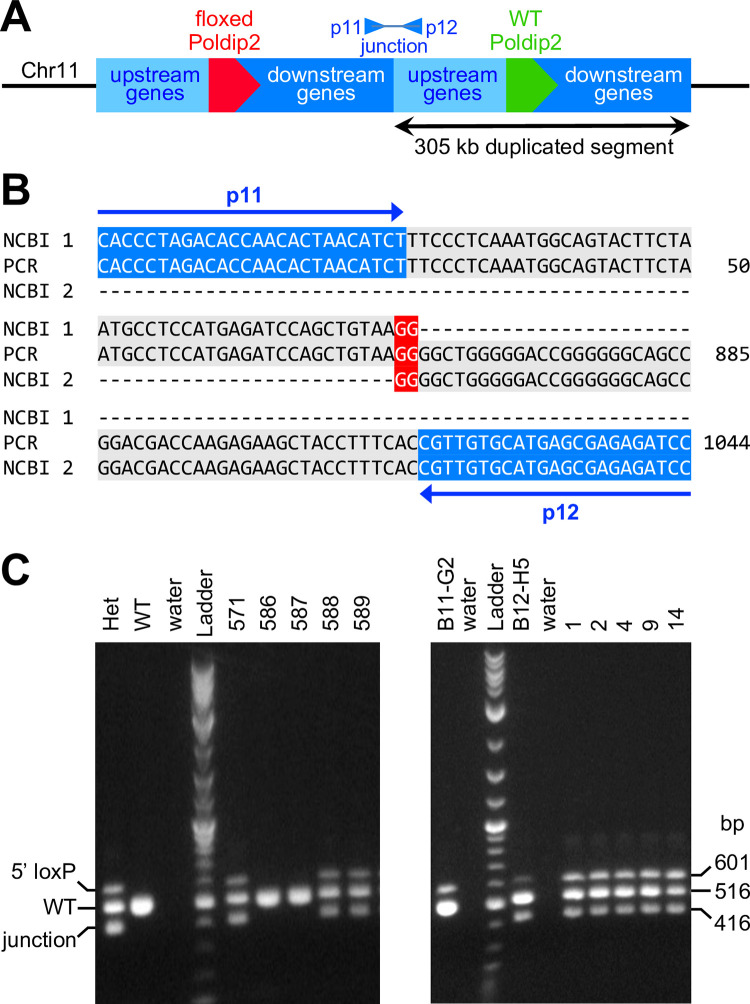
Fig 5. Elucidation of the rearrangement structure provides a new genotyping method.
A: Schematic of a possible genomic rearrangement with both duplicated segments located side by side in the same orientation and connected by a junction. One copy carries floxed Poldip2, while the other retains the WT allele. B: Genomic DNA from a homozygous floxed Poldip2 mouse was amplified by PCR with primers p11 and p12, designed to anneal near the edges of the duplicated segment. A single ~1 kb PCR product was generated, thus confirming the structure proposed in panel A. Following cloning and sequencing of the PCR product, its 1,044 bp sequence was used in a BLAST search of NCBI’s mouse genome. The alignment shows the ends and the center of the PCR product, including primers highlighted in blue and the GG doublet at the junction highlighted in red. Nucleotides 1–862 are 100% identical to a DNA segment (NCBI 1) located downstream of Poldip2, while nucleotides 861–1,044 are 100% identical to a DNA segment (NCBI 2) located upstream of Poldip2. Sequence coordinates from NCBI GRCm38.p6 (5’ end 78,328,894 and 3’ end 78,634,062) provide the exact length of the duplication: 305,169 bp, thus refining the result from Fig 4B. C: A new genotyping method with two primer pairs was designed to detect both the 5’ loxP site (p7 & p8 as in Fig 2) and the duplication junction (p13 & p12) in a single PCR tube. A number of floxed Poldip2 mice were re-genotyped with this new method with the hope of finding floxed animals without duplication. A representative gel (left) shows that the floxed allele and the mutation were always detected together. Furthermore, the duplication was also found in DNA samples from the earliest Het floxed mice of the colony (right gel, mouse numbers are shown at the top) and in the ES cell clone (B12-H5) from which they derived, indicating that the duplication is stable and occurred during gene targeting. However, another ES cell clone (B11-G2) appeared to be free from the same defect.