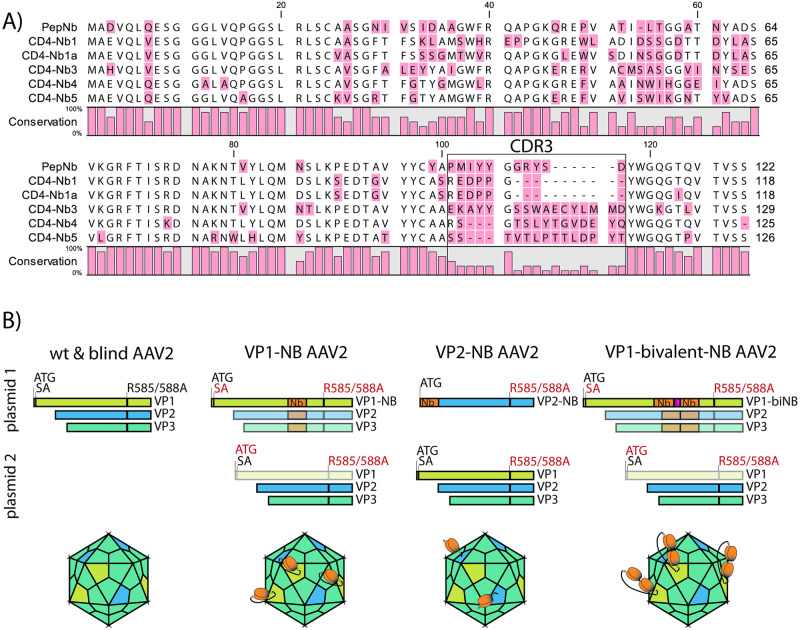
Fig 1. Nanobody (Nb) sequences and AAV2-Nb design.
(A) Amino acid alignment of the Nb sequences used in this study. Nb annotation is in accordance with the study by Traenkle and coworkers [39], which describes thorough Nb characterization and application for in vivo imaging. Of note, CD4-Nb1a is not specified in Traenkle et al., as its complementary determining region 3 (CDR3, black box) is identical to CD4-Nb1 (amino acids 101–117 in the alignment). (B) Plasmid constructs used for production of indicated AAV2 particles and schematic representation of the resulting capsids. Equimolar amounts of plasmids were co-transfected into HEK293T cells (see Methods). For clarity, adenoviral helper plasmid and vector encoding the eGFP reporter included in all co-transfections are not depicted. Essential changes to optimized viral capsid proteins VP1-3 are indicated: red = mutated / black = wt. Disabled open reading frames are greyed out. ATG = start codon; SA = splice acceptor site; Nb = full length nanobody sequence as presented in A (orange); L = glycine-serine linker genetically fusing nanobody monomers in the bivalent constructs (pink); R585A/R588A = Arg to Ala amino acid substitution at residues 585 and 588 (blind). AAV capsids were rendered with help of the browser tool by Antonio Negrón [42].