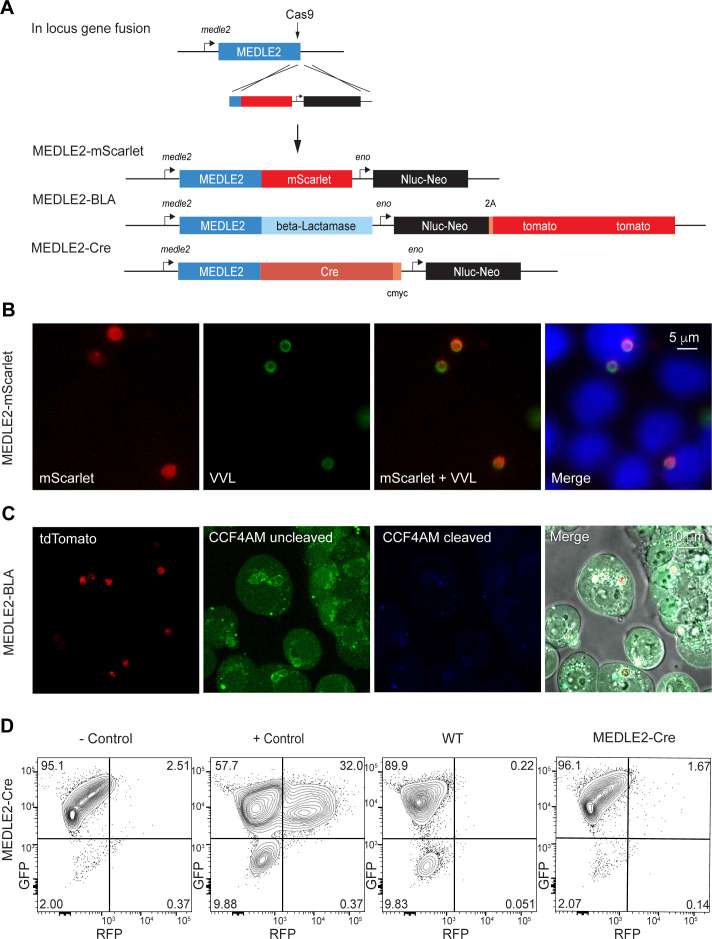
Figure 4. Ordered reporters disrupt MEDLE2 export.
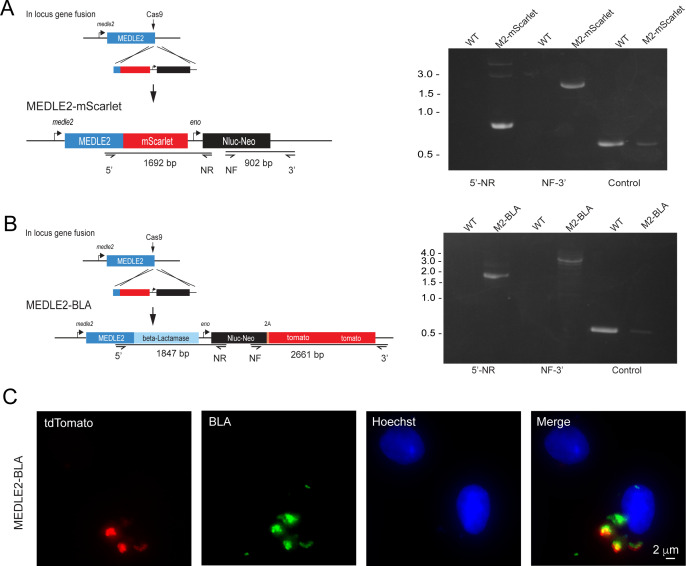
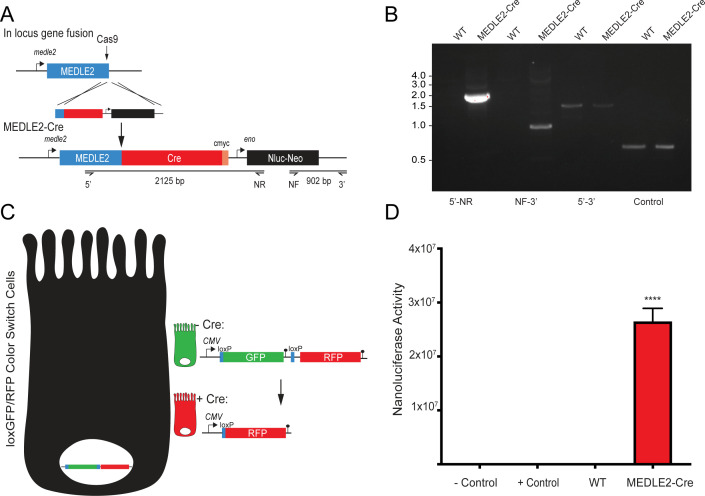
(A) Schematic map of the MEDLE2 locus targeted for insertion of three different reporter genes (mScarlet, beta-lactamase, or Cre recombinase), nanoluciferase (Nluc), and the selection marker (Neo). The guide RNA and flanking sequences used here were the same as those employed to generate MEDLE2-HA transgenic parasites (see Figure 2—figure supplement 1, Figure 4—figure supplement 1 for more detail). (B) MEDLE2-mScarlet parasites were used to infect HCT-8 cells and fixed for immunofluorescence assay (IFA) across a time course. Data shown are from 10 hr post infection, which is representative of the MEDLE2 localization observed at all time points. Red, Medle2-mScarlet; green, parasites (VVL); blue, Hoechst. (C) HCT-8 cells were infected with MEDLE2-BLA C. parvum for 24 hr before incubation with the CCF4-AM beta-lactamase substrate and visualization by live microscopy. This experiment was repeated three times. Red, parasites (tdTomato); green, uncleaved CC4F-AM; blue, cleaved CCF4-AM; gray, DIC. We attribute lack of CCF4-AM cleavage to failure of MEDLE2-BLA to export (Figure 4—figure supplement 1). (D) MEDLE2-Cre parasites were used to infect loxGFP/RFP color switch HCT-8 cells ((Figure 4—figure supplement 2) for schematic representation). After 48 hr, cells were subjected to flow cytometry. Live, single cells were gated based upon forward and side scatter, and green fluorescence (GFP) and red fluorescence (RFP) were measured to detect Cre recombinase activity. Despite robust infection, MEDLE2-Cre-infected cultures did not express RFP (Figure 4—figure supplement 2) compared to the positive control that was transiently transfected to express Cre recombinase.