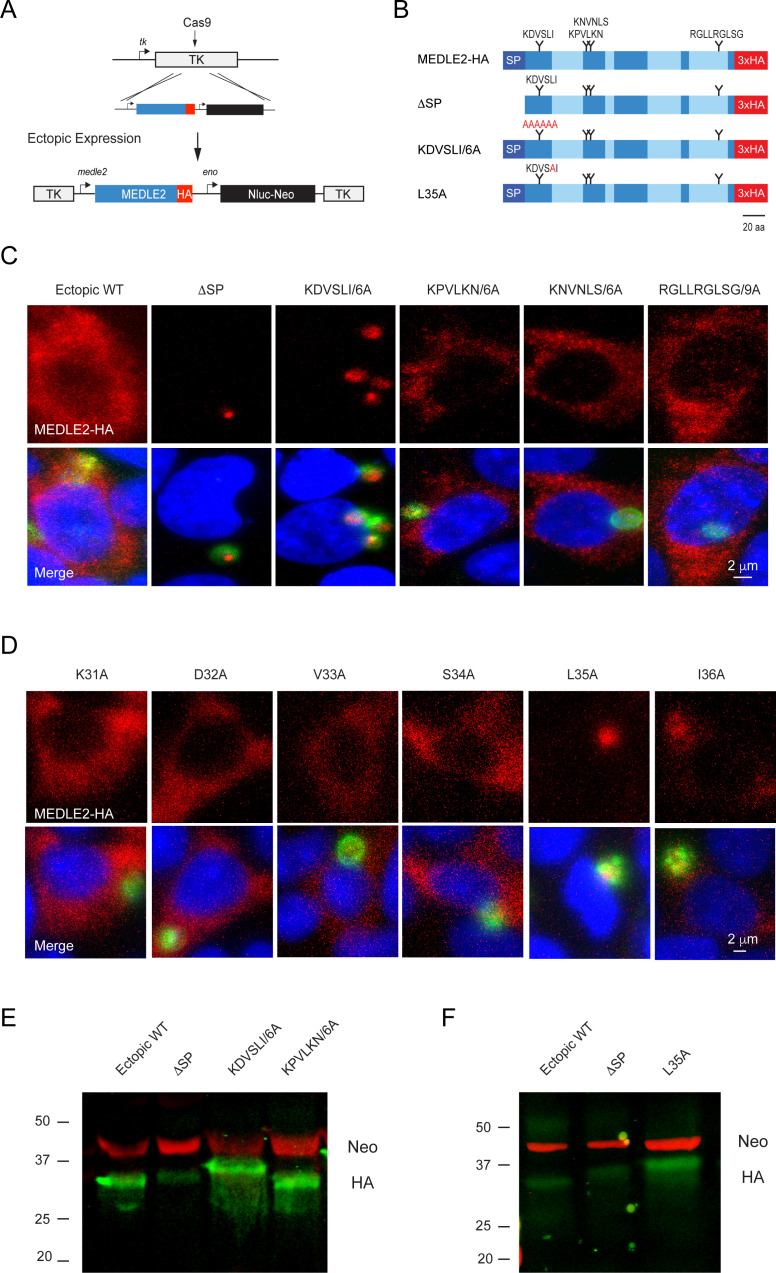
Figure 5. MEDLE2 contains a host-targeting motif that is processed during export.
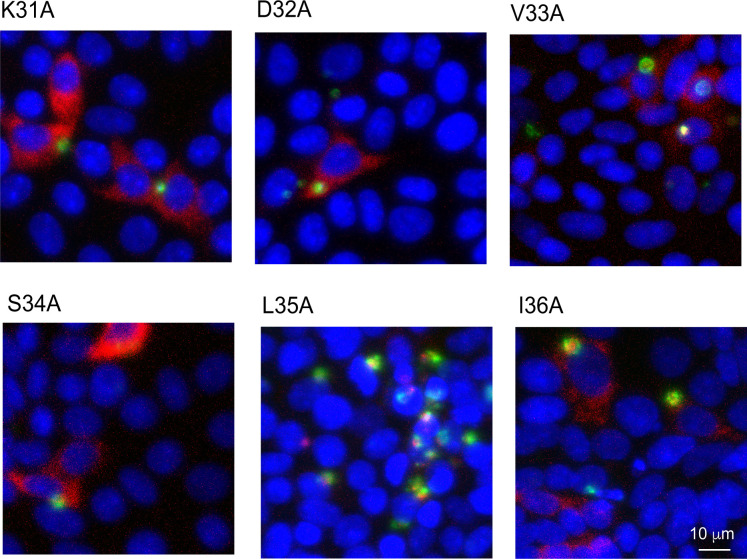
(A) Map showing the strategy used to engineer an ectopic copy of MEDLE2-HA in the thymidine kinase (TK) locus. Expression of an ectopic copy of MEDLE2-HA was driven by the MEDLE2 promoter. All point mutations were confirmed by Sanger sequencing (Figure 5—figure supplement 1). (B) Schematic representation of the MEDLE2 mutants generated using the strategy outlined in (A). The signal peptide (SP) is represented by dark blue, and low-complexity regions are shown in light blue. Candidate motifs targeted for mutagenesis are indicated with black triangles, and mutagenized amino acids are shown in red for two representative mutants. (C, D) Mutant parasites were used to infect HCT-8 cells and fixed for immunofluorescence assay (IFA) after 24 hr. For mutants shown in (C), the entire candidate motif was replaced with a matching number of alanine residues (e.g., KDVSLI/6A → AAAAAA). For mutants shown in (D), each individual amino acid in the KDVSLI sequence was changed to alanine. Red, hemagglutinin (HA)-tagged protein; green, parasites (VVL); blue, Hoechst. We note that SP and leucine 35 within the KDVSLI sequence are required for MEDLE2 export. (E, F) 5 × 106 transgenic oocysts were used to infect HCT-8 cells for 48 hr before preparation of whole-cell lysates. Proteins were separated by for SDS-PAGE and analyzed by western blot. The resulting blots for infections with whole motif mutants (E) and individual amino acid point mutants (F) are shown. Red, neomycin; green, HA. Note that when mutants are expressed in mammalian cells and not C. parvum the resulting proteins do not show any size differences (Figure 5—figure supplement 2).
Figure 5—figure supplement 1. Sanger sequencing confirming the generation of MEDLE2 mutants.
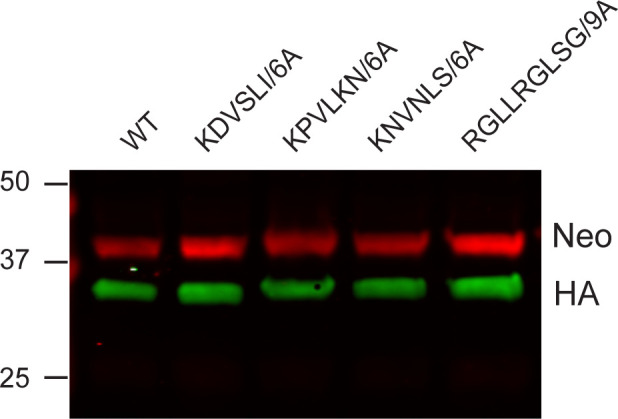
Figure 5—figure supplement 2. MEDLE2 mutants are of the same size as wild type (WT) MEDLE2 when expressed in HEK293T cells.